- (Optional) Creating conda environment
conda create -n lavis python=3.8
conda activate lavis- for development, you may build from source
git clone https://github.com/salesforce/LAVIS.git
cd LAVIS
pip install -e .Raw data are avaliable at https://ftp.uniprot.org/pub/databases/uniprot/previous_releases/release-2023_04/knowledgebase/
The experimental train/val/test set of Swiss-Prot are avaliable at data/swissprot_exp
ESM2 embeddings generation: https://github.com/facebookresearch/esm
The generation command:
git clone https://github.com/facebookresearch/esm.git
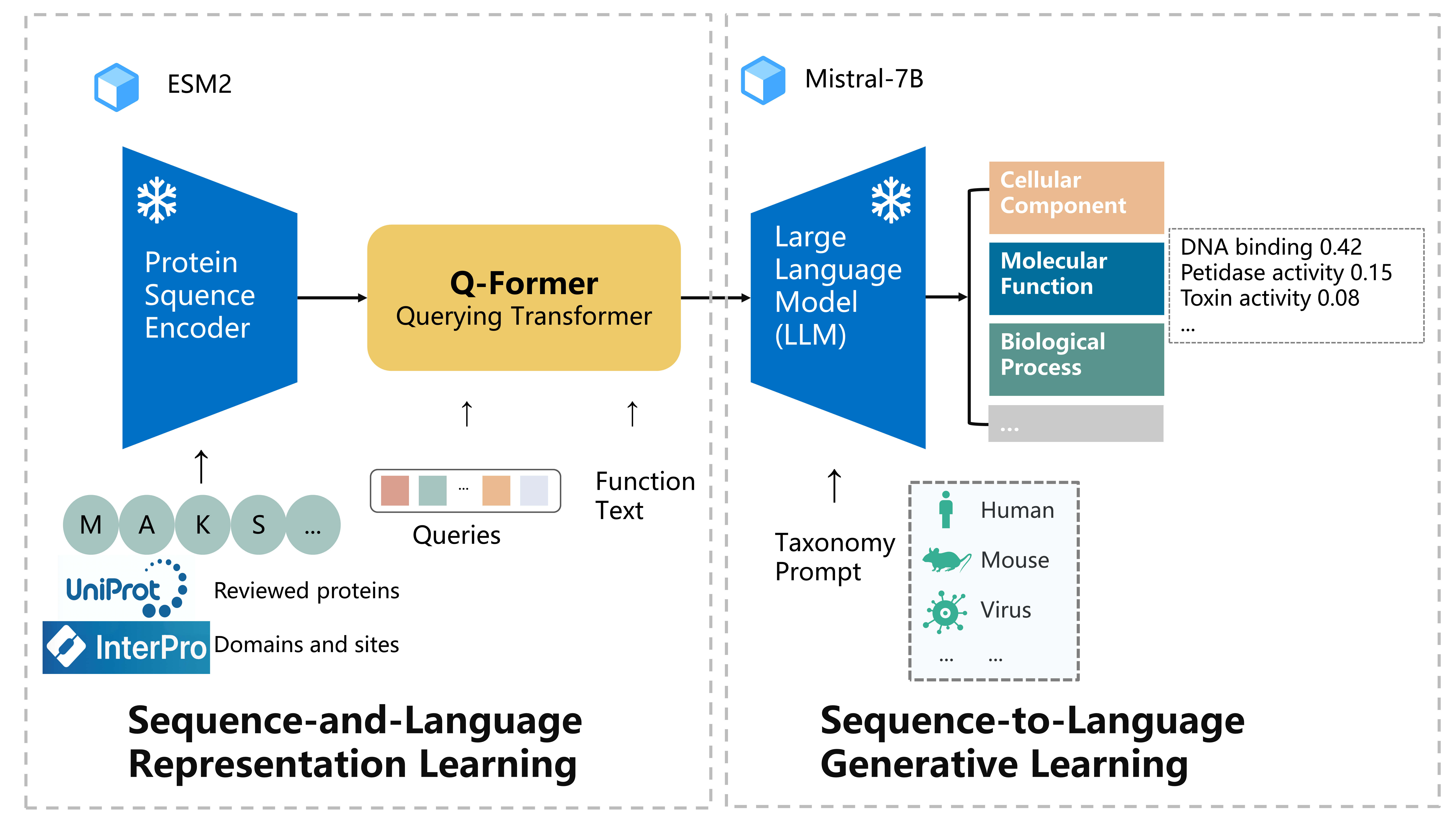
python scripts/extract.py esm2_t33_650M_UR50D you_path/protein.fasta you_path_to_save_embedding_files --repr_layers 33 --truncation_seq_length 1024 --include per_tokdata config: lavis/configs/datasets/protein/GO_defaults_cap.yaml
stage1 config: lavis/projects/blip2/train/protein_pretrain_stage1.yaml
stage1 training command: run_scripts/blip2/train/protein_pretrain_domain_stage1.sh
stage2 config: lavis/projects/blip2/train/protein_pretrain_stage2.yaml
stage2 training/finetuning command: run_scripts/blip2/train/protein_pretrain_domain_stage2.sh
You can download our trained models from drive: https://drive.google.com/drive/folders/1aA0eSYxNw3DvrU5GU1Cu-4q2kIxxAGSE?usp=drive_link
config: lavis/projects/blip2/eval/caption_protein_eval.yaml
command: run_scripts/blip2/eval/eval_cap_protein.sh
We provide an example in FAPM_inference.py. You can change the example protein to you custom case