This code is for our paper:
@article{wang2024open,
title={Open access dataset, code library and benchmarking deep learning approaches for state-of-health estimation of lithium-ion batteries},
author={Wang, Fujin and Zhai, Zhi and Liu, Bingchen and Zheng, Shiyu and Zhao, Zhibin and Chen, Xuefeng},
journal={Journal of Energy Storage},
volume={77},
pages={109884},
year={2024},
publisher={Elsevier}
}
paper link: https://doi.org/10.1016/j.est.2023.109884
Please cite our paper if you find it useful.
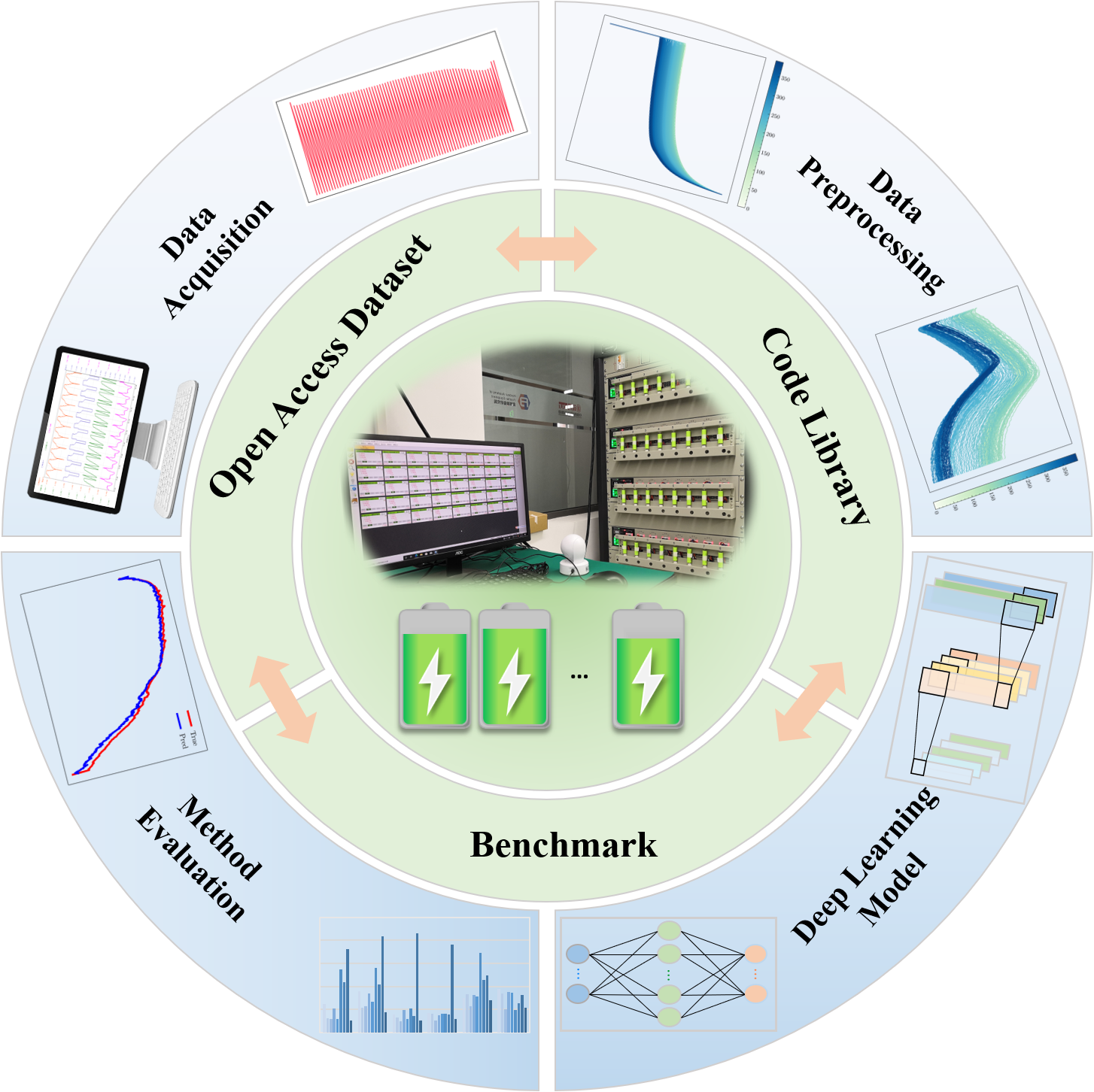
This is a benchmarking code for state-of-health estimation of lithium-ion batteries.
It contains 100 batteries, 5 deep learning models, 3 input types, 3 normalization methods.
You can choose which model to train, which input type, which battery, and which normalization method by changing the following parameters:
--model: choices=['CNN','LSTM','GRU','MLP','Attention']
--data: choices=['XJTU','MIT']
--batch: you can select [1,2,3,4,5,6] for XJTU, and [1,2,...,9] for MIT
--test_battery_id: 1-8 for XJTU (1-15 for batch-2), 1-5 for MIT
--input_type: choices=['charge','partial_charge','handcraft_features']
--normalized_type: choices=['minmax','standard']
# if you select 'minmax', you can set:
--minmax_range: choices=[(0,1),(-1,1)]for example:
python main.py --model CNN --data XJTU --batch 1 --test_battery_id 1 --input_type handcraft_features --normalized_type minmax
You can choose how many times to train.
The results of each training will be saved in the results folder.
We only provide a baseline here, and you can make improvements based on it, such as using better models, better hyperparameters, better training strategies, etc.
The data contained in the data folder has been preprocessed and can be directly used as input for the aforementioned 5 models.
The raw data can be found at: XJTU battery dataset.
It includes detailed descriptions along with some codes for feature extraction.