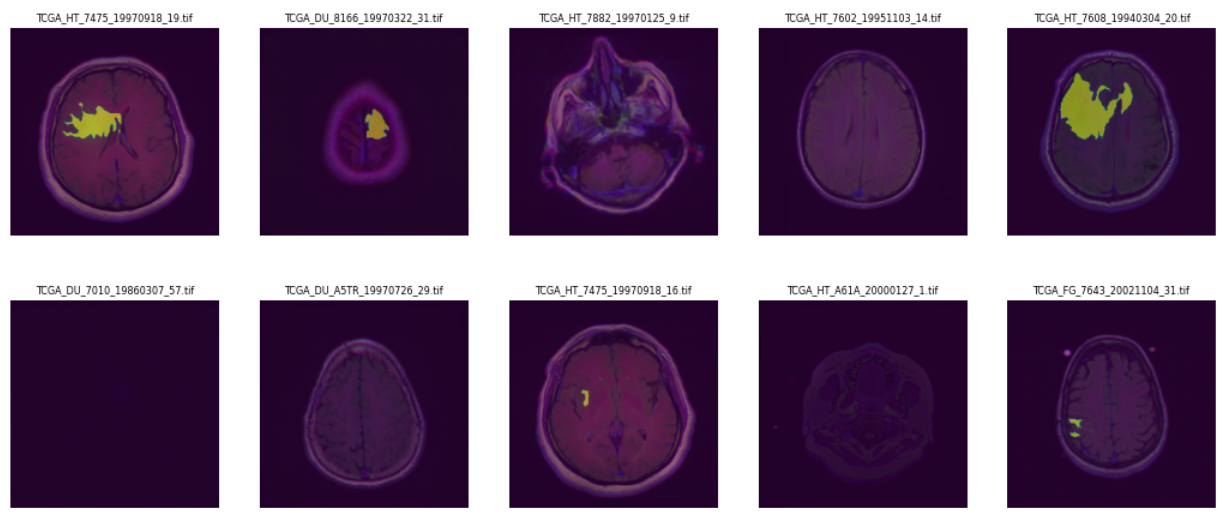
This repository implements two semantic segmentation models UNet-3 (3 skip-cons) and UNet-4 (4 skip cons). the models can be found in ./imgseg/networks.py
The documentation of the whole procedure from start to finish is
documented in the notebook documentation.ipynb.
The models can be downloaded from here: https://drive.google.com/drive/folders/1nm74FqZckW0pwa-L1O_SwnRV7-iAA-sR?usp=sharing
During the evaluation the segmentation model with the name dice_unet4_50eps_16bs.pth performed best and it is recommended to download this model for inference. The evaluation was made on certain patients from the used kaggle dataset for training. I can not guarantee that the model performs good on other images, which might origin from a different distribution.
- Unet with 4 skip connections
- Trained with normal Dice-Loss
- 50 epochs in total
- Batch size of 16
- Adam optimizer with additional step-wise learning rate scheduler
- Clone project locally via SSH:
git clone git@github.com:SimonStaehli/image-segmentation.gitor via HTTPS:
git clone https://github.com/SimonStaehli/image-segmentation.git-
Download models from Google Drive and add it to the folder
./modelshere: https://drive.google.com/drive/folders/1nm74FqZckW0pwa-L1O_SwnRV7-iAA-sR?usp=sharing -
Install required packages
pip install -r requirements.txtNote: if there is a problem with the installation please refer to the official pytorch installation guide: https://pytorch.org/get-started/locally/
docker build . -f Dockerfile -t brain-mri-semantic-segmentationdocker run -it brain-mri-semantic-segmentationRequires all installation steps already made.
Following command will segment all images in the folder image_in and store the masks in the folder image_out.
python inference.py -i image_in -o image_out --overlay TrueDataset used for training: https://www.kaggle.com/datasets/mateuszbuda/lgg-mri-segmentation
Kaggle Notebook used for training: https://www.kaggle.com/code/simonstaehli/image-segmentation