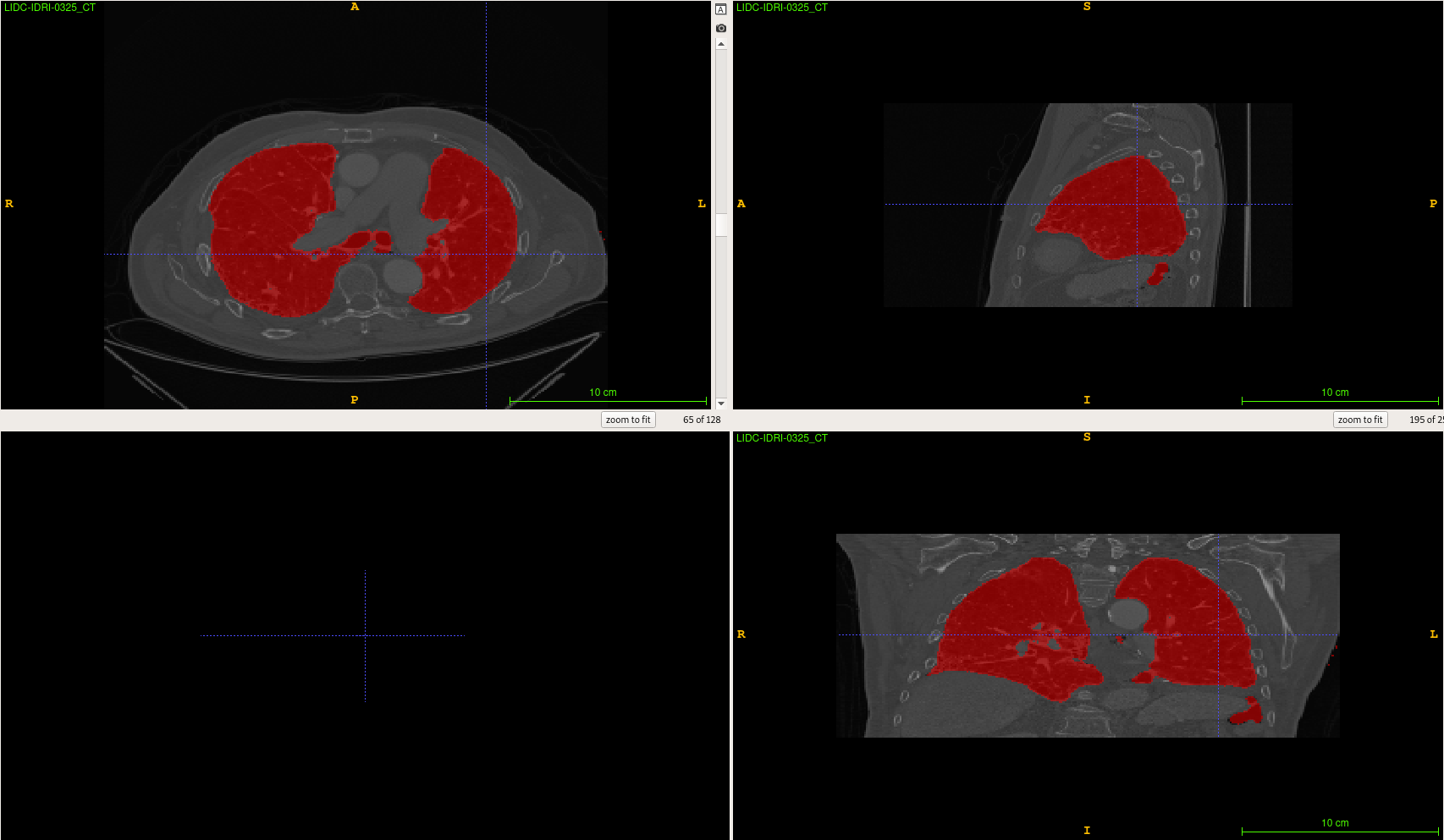
Lung Segmentation
Lung Segmentation using a U-Net model on 3D CT scans.
Current results example :
Getting started
Installation
Our base wmlce conda environment does not come with SimpleITK nor pynrrd, two required python libraries to run this code.
- To install
pynrrd:
$ pip install pynrrd
- To install
SimpleITK(from.whl):
$ pip install SimpleITK-1.2.0+gd6026-cp36-cp36m-linux_ppc64le.whl
- If you do not have access to the whl file, you need to build it (on power pc)
Tree
.
+-- data/
+-- dataset.py : Class describing the dataset we use for lung segmentation
+-- utils.py : Script for manipulating medical files
+-- config.json
+-- eval.py
+-- model.py : U-Net model definition
+-- predict.py : Inference script to run infer lung mask on a CT-scan
+-- README.md : This documentation file
+-- train.py : Train script to train a new lung segmentation model
Data
The data used is the TCIA LIDC-IDRI dataset Standardized representation (download here), combined with matching lung masks from LUNA16 (not all CT-scans have their lung masks in LUNA16 so we need the list of segmented ones).
3 parameters have to be fulfilled to use available data:
labelled-list: path to thepicklefile containing the list of CT-scans from the TCIA LIDC-IDRI dataset for which we have access to the lung segmentation masks through the LUNA16 dataset.scans: path to the TCIA LIDC-IDRI dataset.masks: path to the LUNA16 dataset containing lung masks.
You can manipulate data trough the data/dataset.py (class describing our lung segmentation dataset) and data/utils.py (tools for manipulating medical files) files.
Predictions
To perform predictions on unseen CT-scans, run for example (wmlce on powerai):
$ data=/wmlce/data/projects/jfr/data/LIDC-IDRI/LIDC-IDRI-0325/1.3.6.1.4.1.14519.5.2.1.6279.6001.815399168774050638734383723372/1.3.6.1.4.1.14519.5.2.1.6279.6001.725023183844147505748475581290/LIDC-IDRI-0325_CT.nrrd
$ output_path=/wmlce/data/projects/jfr/ls_output
$ nb_classes=1
$ start_filters=32
$ python3 predict.py -d $data -o $output_path -c $nb_classes -f $start_filters -t [-e]
- See
python3 predict.py --helpfor more information.
Evaluation
To perform evaluation using the existing model, run for example (wmlce on powerai):
$ LABELLED_LIST=/wmlce/data/retina-unet/data/labelled.pickle
$ MASKS=/wmlce/data/retina-unet/data/lung_masks_LUNA16
$ SCANS=/wmlce/data/retina-unet/data/LIDC-IDRI
$ NB_CLASSES=1
$ START_FILTERS=32
$ python3 eval.py --labelled-list $LABELLED_LIST --masks $MASKS --scans $SCANS --nb-classes $NB_CLASSES --start-filters $START_FILTERS
- See
python3 eval.py --helpfor more information.