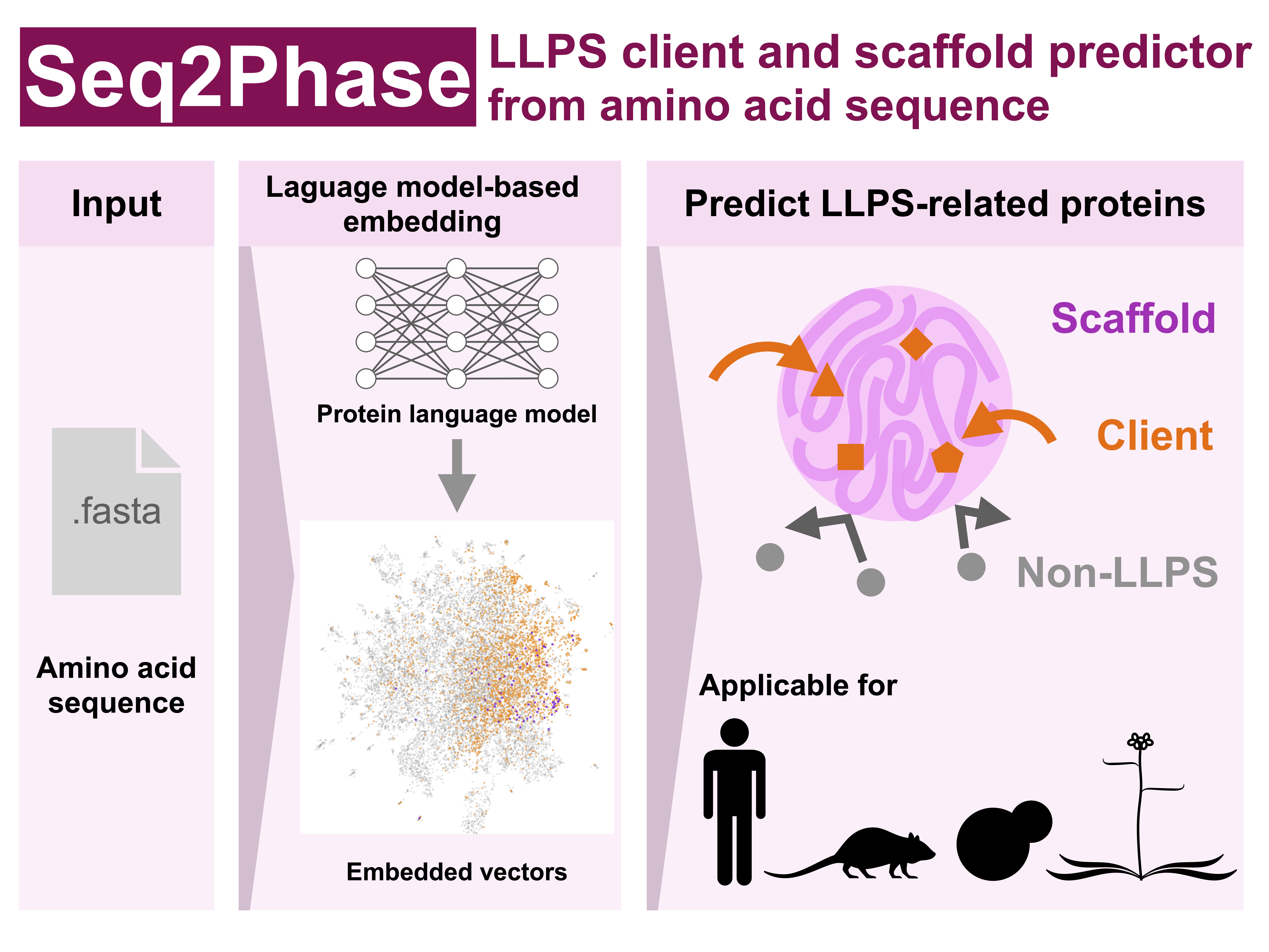
LLPS-related protein predictor from amino acid sequence
- Purpose: Predicts LLPS client proteins from amino acid sequences.
- Unique Focus: Concentrates on client proteins in LLPS, which are proteins localizing in condensates but not essential for their formation.
- Technology: Utilizes large language model (LLM) embeddings and a range of machine learning (ML) models (SVM, RF, HGBC, NN).
- Application: Capable of proteome-wide analysis.
- Utility: Aids in identifying client protein candidates for deeper biological research.
- Broad Applicability: Effective across multiple species including humans, mice, yeast, and Arabidopsis thaliana.
- Limitations:
- Not tested in prokaryotes.
- Computations can be slow when run only on a CPU. For CPU usage, the
--less_dataoption is recommended.
- Function: Facilitates intracellular compartmentalization without biological membranes.
- Process: Forms membraneless organelles like nucleoli and p-bodies.
- Components: Involves two types of proteins - scaffolds and clients.
- Scaffolds: Essential for condensate formation.
- Clients: Localize within condensates, performing various roles but not required for condensate formation.
- Relevance: Linked to cellular processes and various diseases, including neurodegenerative disorders.
Install Bio Embeddings for protein embedding using LLM and ML
pip install bio-embeddings==0.2.2
Install PyTorch (<=1.10.0,>=1.8.0) suitable for your machine
Linux and Windows
#CUDA 11.1
pip install torch==1.10.0+cu111 torchvision==0.11.0+cu111 torchaudio==0.10.0 -f https://download.pytorch.org/whl/torch_stable.html
#CPU only
pip install torch==1.10.0+cpu torchvision==0.11.0+cpu torchaudio==0.10.0 -f https://download.pytorch.org/whl/torch_stable.html
MacOS
pip install torch==1.10.0 torchvision==0.11.0 torchaudio==0.10.0
More information: PyTorch
Run train_model.py first, then seq2phase.py.
python train_model.py
python seq2phase.py -i input.fasta -o output.tsv- Initial Setup: Run
train_model.pyonce before usingseq2phase.py. Once executed,seq2phase.pycan be used multiple times. - For CPU Environments: Using the
--less_dataoption is recomended.
- Input: The script takes an input file in FASTA format.
- Output: Generates an output file containing client and scaffold scores.
- Interpreting Scores: If both scores of a protein are high (>0.5), it is considered a scaffold protein.
Kazuki Miyata, Wataru Iwasaki, Seq2Phase: Language Model-based Accurate Prediction of Client Proteins in Liquid-Liquid Phase Separation, Bioinformatics Advances, 2023;, vbad189.
https://doi.org/10.1093/bioadv/vbad189
Kazuki Miyata (The University of Tokyo) miyatakazuki381@g.ecc.u-tokyo.ac.jp