biolgca is a Python package for simulating different types of lattice-gas
cellular automata (LGCA) in the biological context.
LGCA are a subclass of cellular automata with an extended state space that allows each particle/cell to have a direction. For a more detailed introduction see the Wikipedia article. They present a mesoscopic modelling framework to analyse collective phenomena, e.g. cell migration. Use cases are demonstrated in this paper.
The package is intended for use in ongoing research as well as to exemplify the unique advantages of the framework. It is under active development. The types of LGCA currently supported are:
- classical LGCA (volume exclusion, all particles/cells have the same properties)
- identity-based LGCA (volume exclusion, particles/cells can have individual properties)
- classical LGCA without volume exclusion (all particles/cells have the same properties)
- identity-based LGCA without volume exclusion (particles/cells can have individual properties)
These can be simulated in a 1D, 2D square or 2D hexagonal lattice. A library of interaction rules (documentation under construction) is already implemented. Adding a custom interaction rule or customising other parts of the simulation (e.g. the interaction radius) is easy.
Current analysis possibilities include plots of:
- density (+ animation)
- flux (+ animation)
- flow (+ animation)
- full state space (+ animation)
- scalar field
- vector field
The internal state of the LGCA is always accessible for computational analysis.
from lgca import get_lgca
# specify the alignment interaction
# LGCA geometry: hexagonal lattice, no resting channels and reflecting boundary conditions
# nodes will be initialised randomly
lgca = get_lgca(geometry='hex', interaction='alignment', bc='refl')
# simulate for 132 timesteps without viewing the result
# record the full channel configuration of the lattice at all timesteps
lgca.timeevo(timesteps=132, record=True)
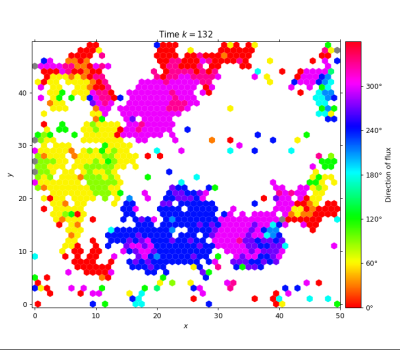
# plot the flux to see clusters of aligned particles moving in the same direction
lgca.plot_flux()from lgca import get_lgca
# specify the excitable medium interaction with interaction parameter N=20
# LGCA geometry: 20 resting channels and reflecting boundary conditions
lgca = get_lgca(interaction='excitable_medium', restchannels=20, N=20, bc='refl')
# initialise a custom lattice configuration
lgca.nodes[...] = 0
lgca.nodes[:lgca.lx // 2, :, :lgca.velocitychannels] = 1 # 6 moving particles per node in the left half of the lattice
lgca.nodes[:, :lgca.ly // 2,
lgca.velocitychannels:] = 1 # 6 resting particles per node in the lower half of the lattice
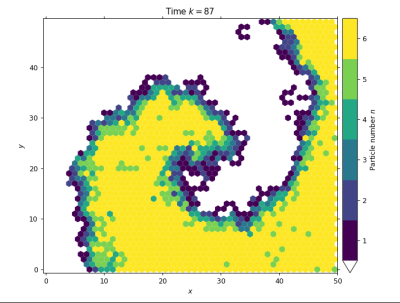
# view a live simulation: density profile of the velocity channels only
lgca.live_animate_density(channels=slice(0, lgca.velocitychannels), vmax=lgca.velocitychannels)
# the following image results from stopping the simulation after k=87 timestepsfrom lgca import get_lgca
# identity-based LGCA: each cell can have own properties
# specify the "Go and grow" interaction: cancer cells proliferate and migrate,
# where the birth rate mutates
# LGCA geometry: 1D lattice, 6 resting channels
lgca = get_lgca(interaction='go_and_grow', ib=True, geometry='lin', restchannels=6)
# initialise a custom lattice configuration
lgca.nodes[...] = 0
lgca.update_dynamic_fields()
lgca.nodes[lgca.dims[0] // 2, :] = 1 # 1 fully filled node at the center
# simulate for 200 timesteps and record the full lattice configuration
lgca.timeevo(timesteps=200, record=True)
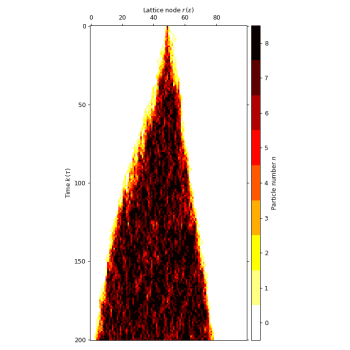
# plot cell density to see the tumour growth over time
lgca.plot_density(colorbarwidth=0.2) # plot on the left
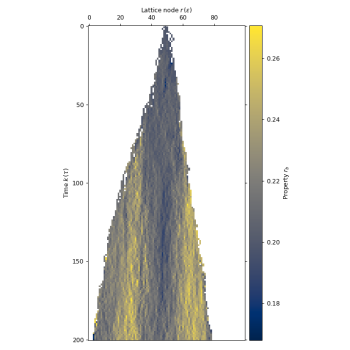
# plot the birth rate to inspect its increase
lgca.plot_prop_spatial(propname='r_b') # plot on the rightbiolgca heavily depends on numpy and matplotlib. To install all dependencies
for using the package, run this from a command line:
pip install matplotlib==3.3.2 numpy scipy sympy(On Windows, a terminal that understands pip out of the box can be opened in Anaconda in the
"Environments" tab, clicking on the triangle next to the environment's name.)
If you want to add code to the package, in order to run tests and build documentation you should also install:
pip install pytest==6.2.5 Sphinx==4.4.0 sphinx-autodoc-typehints alabaster numpydocbiolgca does not have a package distribution yet. To use it, clone (or unzip the download of)
the master branch of the repository into a folder of your choice.
If your scripts are not going to be in the biolgca folder, add the following to
the beginning of the Python files:
import sys
sys.path.insert(1, "/absolute/path/to/folder/biolgca")To use the package, simply import the get_lgca function:
from lgca import get_lgcaIt will return an instance of the correct class of LGCA according to the passed
arguments, e.g. a 1D identity-based LGCA (IBLGCA_1D), already initialised with
the specified initial conditions. This can be used to simulate
the automaton with the specified interaction and inspect the results.
# request the LGCA
lgca = get_lgca(ib=True, geometry='1d', interaction='random_walk')
# simulate for 50 timesteps
lgca.timeevo(timesteps=50)
# plot the development of the particle/cell density over time
lgca.plot_density()The Tutorial guides you through the argument options.
The structure of the package and its functionalities are detailed in the documentation. There you will also find coding examples (under construction).
Issues are tracked on the GitHub page. We collect both bugs and feature ideas there.
For guidelines how to add code to and maintain the repo visit the Wiki.
For remaining questions you can contact us via E-mail.
Simon Syga: simon.syga@tu-dresden.de
Bianca Güttner: bianca.guettner@nct-dresden.de
BSD 3-clause license (see LICENSE file or online resource).
Copyright (C) 2018-2022 Technische Universität Dresden, contact: simon.syga@tu-dresden.de.