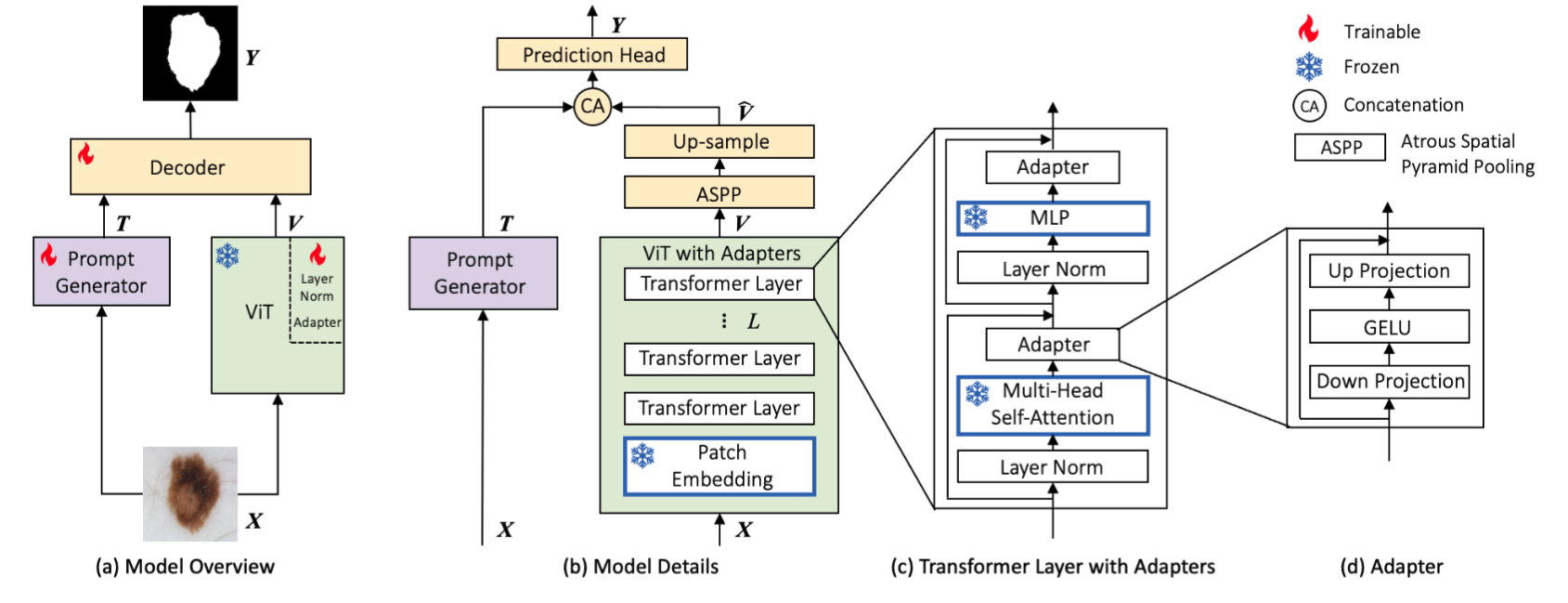
Architecture of AViT: (a) Model overview with its prompt generator (a shallow CNN network), a large pre-trained ViT backbone with adapters, and a compact decoder. (b) Model details. (c) Details of a transformer layer with adapters. (d) Details of our adapters. During training, all modules in (b,c,d) contoured with blue borders are frozen, which encompasses 86.3% of AViT's parameters.
This is a PyTorch implementation for AViT: Adapting Vision Transformers for Small Skin Lesion Segmentation Datasets, MICCAI ISIC Workshop 2023.
We also include plenty of comparing models in this repository: SwinUnet, UNETR, UTNet, MedFormer, SwinUNETR, H2Former, FAT-Net, TransFuse, AdaptFormer, and VPT (Please go to the paper to find the detailed information of these models).
If you use this code in your research, please consider citing:
@inproceedings{du2023avit,
title={{AViT}: Adapting Vision Transformers for Small Skin Lesion Segmentation Datasets},
author={Du, Siyi and Bayasi, Nourhan and Hamarneh, Ghassan and Garbi, Rafeef},
booktitle={26th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2023) ISIC Workshp},
This code is implemented using Python 3.8.1, PyTorch v1.8.0, CUDA 11.1 and CuDNN 7.
conda create -n skinlesion python=3.8
conda activate skinlesion # activate the environment and install all dependencies
cd MDViT/
conda install pytorch==1.11.0 torchvision==0.12.0 torchaudio==0.11.0 cudatoolkit=11.3 -c pytorch
# or go to https://pytorch.org/get-started/previous-versions/ to find a right command to install pytorch
pip install -r requirements.txt- Please run the following command to resize original images into the same dimension (512,512) and then convert and store them as .npy files.
python Datasets/process_resize.py- Use Datasets/create_meta.ipynb to create the csv files for each dataset.
- DeiT
- ViT
- Swin Transformer
- ResNet You can also use timm python package to get the pretrained weights of DeiT, ViT, and Swin Transformer and Torchvision python package to get the pretrained weights of ResNet.
- AViT
# ViTSeg_CNNprompt_adapt, SwinSeg_CNNprompt_adapt, DeiTSeg_CNNprompt_adapt
python -u multi_train_adapt.py --exp_name test --config_yml Configs/multi_train_local.yml --model ViTSeg_CNNprompt_adapt --batch_size 16 --adapt_method MLP --num_domains 1 --dataset isic2018 --k_fold 0- BASE
# ViTSeg, SwinSeg, DeiTSeg
python -u multi_train_adapt.py --exp_name test --config_yml Configs/multi_train_local.yml --model ViTSeg --batch_size 16 --adapt_method False --num_domains 1 --dataset isic2018 --k_fold 0- Other skin lesion segmentation models
# SwinUnet, UNETR, UTNet, MedFormer, SwinUNETR, H2Former, FAT-Net, TransFuse
python -u multi_train_adapt.py --exp_name test --config_yml Configs/multi_train_local.yml --model FATNET --batch_size 16 --dataset isic2018 --k_fold 0Please refer to this repository to run BAT.
- AdaptFormer and VPT
# AdaptFormer
python -u multi_train_adapt.py --exp_name test --config_yml Configs/multi_train_local.yml --model AdaptFormer --batch_size 16 --adapt_method AdaptFormer --num_domains 1 --dataset isic2018 --k_fold 0
# VPT
python -u multi_train_adapt.py --exp_name test --config_yml Configs/multi_train_local.yml --model VPT --batch_size 16 --adapt_method False --num_domains 1 --dataset isic2018 --k_fold 0We release the weights for AViT on ViT-Base. play.ipynb showed an example that tests AViT (ViT-B based) on the 4th folder of the ISIC dataset.
| Dataset | ISIC | DMF | SCD | PH2 |
|---|---|---|---|---|
| Weights | Google Drive | Google Drive | Google Drive | Google Drive |