A crucial function of coronavirus proteins is their interaction with RNA. Computational methods can be used to build 3D models of this interaction. But they are static workflows that require all relevant scientific data to be specified up-front. For COVID-19, however, new scientific results come in every day. This requires a new kind of workflow: one that can be modified on the fly with new scientific data, and where new computations and visualizations can be added and adapted, interactively.
SARS-CoV-2 and related coronaviruses are RNA viruses, and proteins interacting with RNA is crucial to their function. In particular, the Nsp9 protein is involved in viral genomic RNA reproduction and interacts with single-stranded RNA. Understanding the details of this interaction may help us to understand how SARS-CoV-2 works and to design inhibitors of its reproduction, helping in the fight against COVID-19.
It is difficult to obtain the 3D coordinates of a protein-RNA molecular complex by experiment. However, a 3D model can be predicted computationally from protein and RNA separately, which is called molecular docking. In molecular docking, it is possible to incorporate scientific data on the interaction (data-driven docking) into the predictions. ATTRACT is an engine for molecular docking that includes support for protein-RNA complexes and data-driven docking. It includes many parameters that can be optimized to obtain the best possible docking result. ATTRACT’s web interface generates a static pipeline for docking two molecules, essentially a shell script, that has to be run by the user on a machine with an installed ATTRACT docking engine. Currently, ATTRACT pipelines are rigid: modifications of the pipeline cannot be made without first interrupting its work, often with loss of results. Consequently, the occurrence of new information requires the replacement of the current pipeline, which is time-consuming and inconvenient.
With the current pandemic, COVID-19 is a very active topic of scientific research. Every day, new research results on COVID-19 and the SARS-CoV-2 virus are published. This makes the docking of Nsp9-RNA a moving target. At any time, scientists can collect the best available scientific data, and create an optimal docking workflow. However, when new data comes in, the new optimal docking workflow may be very different. This requires a more agile kind of workflow: a workflow that the scientific community, in a reproducible and collaborative manner, can easily adapt.
The project aims to create such a workflow. More specifically:
-
Scientific aspect. To collect the current scientific information (literature, predictions) regarding the Nsp9-RNA interaction, and create 3D models of this interaction.
-
Workflow aspect. To design and implement a data-driven docking workflow that optimally takes into account this information. The static pipelines, created by the ATTRACT’s web-interface are taken as a baseline, which will be further adapted.
-
Agility aspect. To allow users to track workflow execution in real-time, customize and adapt it to new scientific results. This consists of two parts. On the one hand, a web interface will be created to modify the input data, aimed at users with no programming experience, primarily biologists.
Interaction with the web interface includes the following actions:
- changing the structure of one or both constituents;
- changing the initial position of the structures (for the docking of 1 RNA structure);
- changing the restraints mode;
- updating information about the binding site of the protein;
- updating information interface and/or contact points of both protein and ssRNA.
In addition, experts in molecular docking will be able to redesign and reprogram any part of the workflow. Both the web interface and the workflow reprogramming are interactive and collaborative: multiple users can make modifications simultaneously, and while being modified, the workflow will continue to run while being modified.
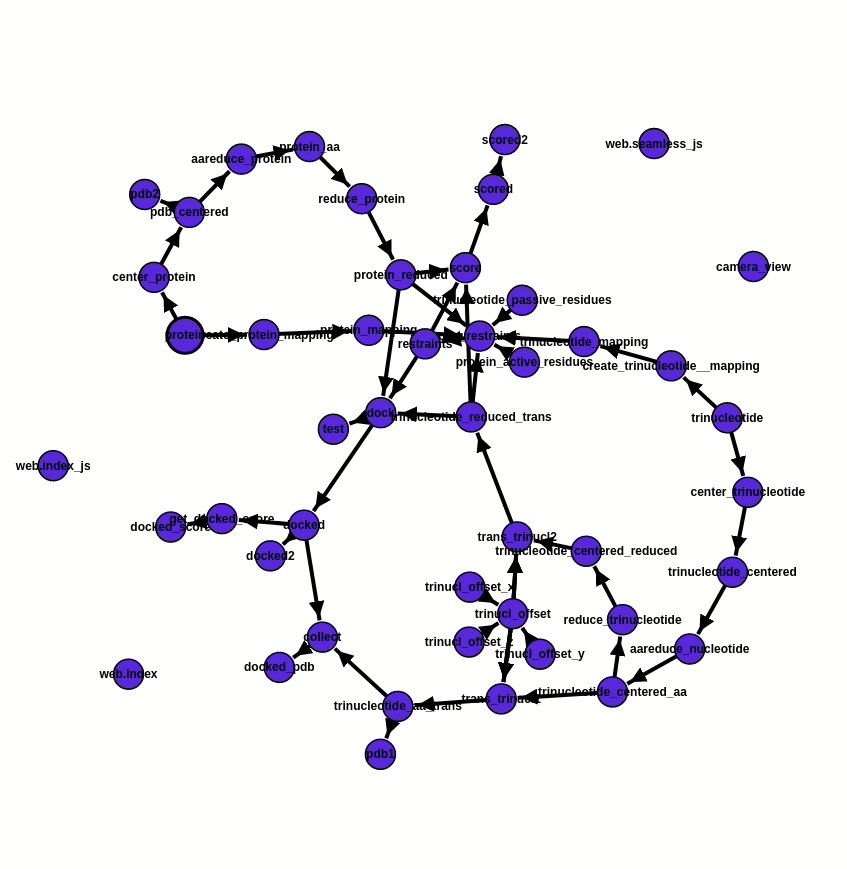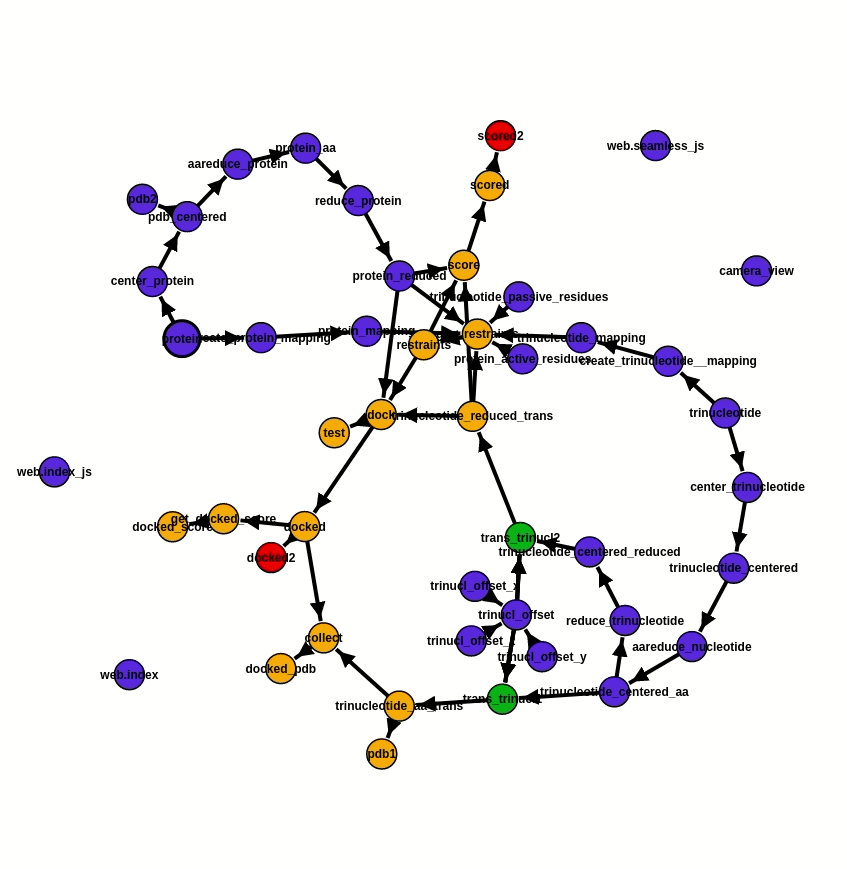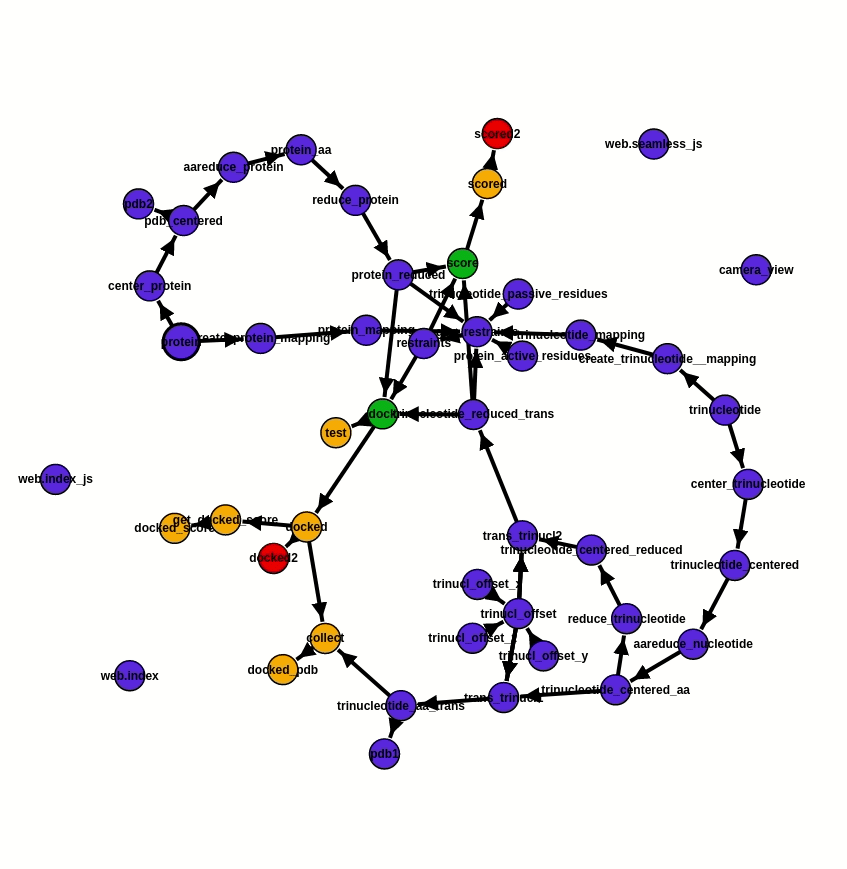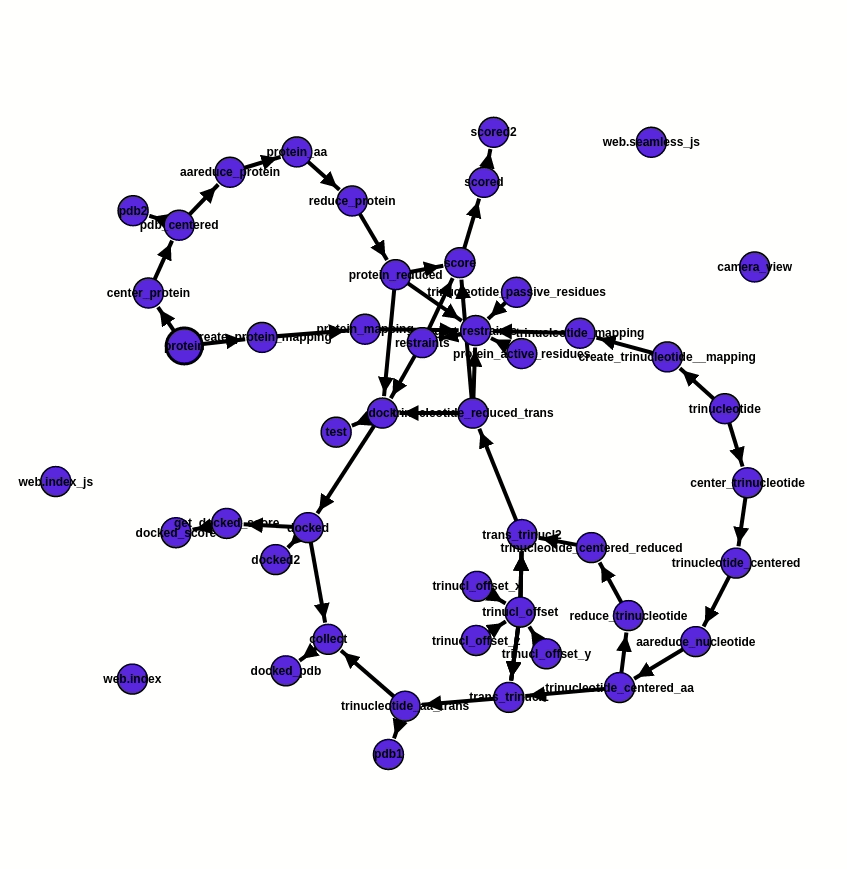
As the foundation of the project, ATTRACT pipeline components are integrated into a workflow via the programming framework Seamless. In Seamless, the workflow is continuously executed while being defined. Data, source code and web interface are all part of the same workflow and can all be changed on the fly. Seamless also provides pipeline visualisation, a Status graph (fig. 2), which allows tracking pipeline execution in real-time. Each pipeline component is presented as a cell, individual bubble, which requires a certain input and provides output after the execution of programmed actions. Execution of action commences automatically as soon as input info is available or updated. In addition, Status graph provides a graphic illustration of the overall progress of the project.
In Seamless, workflows are (re)designed interactively using an IPython console. The Seamless cells of the workflow (data and source code) are mounted to text files that are edited. During work sessions, both IPython console and text files are shared via Visual Studio Code Live Share for real-time collaborative programming.
Within the project, two types of meetings are held on a regular basis:
- Weekly work sessions, structured as listed below:
- newly obtained by participants results are presented and discussed;
- new tasks are formulated and shared between participants;
- a quick recap of the progress since the last session is done;
- collaborative work on the project (1-1.5 hours);
- extra tasks for the next meeting are assigned if any;
- questions about the project are addressed if any.
- Monthly progress reviews: an overview of the project, discussion of the long term aspects.
Weekly meetings are held on Thursdays, at 2 pm, they last approximately 2-3h. A date for each monthly meeting is selected at the beginning of the month.
Currently, several working branches are formed: scientific, back-end development, front-end web development and management.
The scientific part mostly requires biological knowledge, the main tasks are to:
- keep track of relevant information published in articles and reports;
- analyze and interpret available information;
- make sure this information is incorporated in the pipeline (together with participants, responsible for the back-end and front-end part).
The pipeline development requires some biological knowledge (molecular docking, ATTRACT) and programming skills (Python, bash, Seamless) the main tasks are to develop, integrate and test components for the pipeline and the pipeline itself.
The front-end development requires web-development skills (HTML, CSS, JavaScript), the main tasks are to:
- develop a web interface for the created back-end (together with participants, responsible for the back-end);
- make sure it’s user-friendly and accessible for people with little programming experience (together with participants, responsible for the scientific part);
- make sure it’s easily modifiable and allows collaborative work.
The management part consists of keeping track of progress, making sure tasks are shared between participants and can be done by any of the participants.
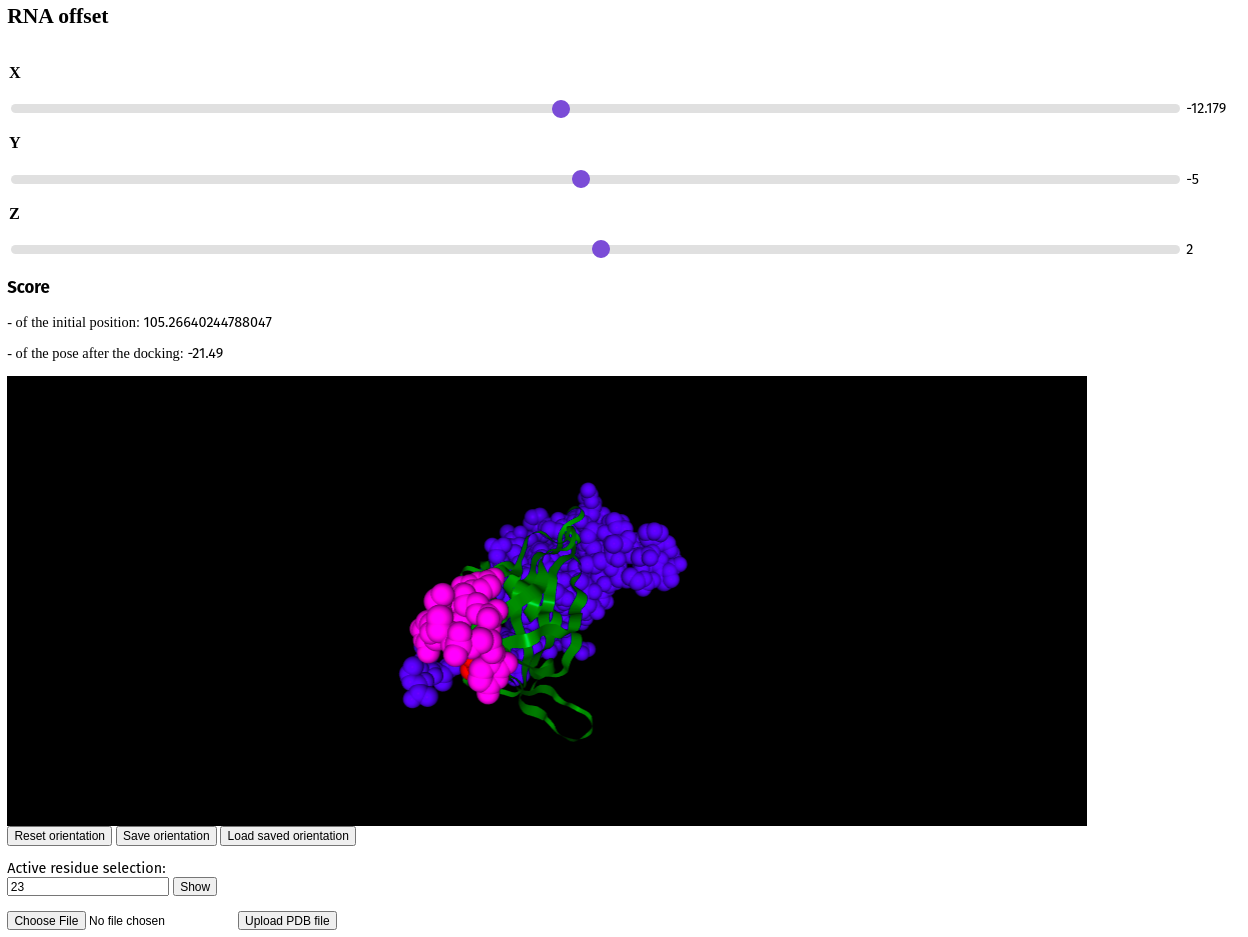
Fig. 1 - The current state of the web interface
Fig. 2 - Status graph of the project from the 20.07.2020. Pipeline execution is reflected by the change of colour. The blue cells are not affected by the updated information, the green ones are the running cells, orange - awaiting for the input, and red - empty or have an error. Here the RNA offset was changed. An update of values occurs immediately, due to which cells trinucl_offset_x/y/z and trinucl_offset do not change their colour on the graph and one can see the changes only in trinucl_offset and trinucl_offset2.
We have come quite far in our development at the moment. 7 weekly meetings have been held and significant progress was made. The preliminary pipeline is ready. The current state of the project allows users to collaboratively view the structure of a molecule, which can be moved (shifted, rotated, zoomed). An arbitrary number of the active residues of the protein binding site can be selected by one of the users and others are able to see these actions. After each alteration docking for current molecules (protein and 1 structure of ssRNA) is done automatically, and users are able to see the resulting 3D structure and its score.
Here is a list of the current achievement:
- In the scientific branch information was collected about:
- conserved residues Arg-10, Lys-52, Trp-53, Arg-55, Arg-74, Phe-75, Lys-86, Tyr-87, Phe-90, Lys-92, Arg-99, and Arg-111 (source);
- conserved residues F73, L86, F88, I95, G98, and G102 (source), where F73, I95, and G98 are found to be critical for stabilizing the dimeric conformation of nsp9 and for nsp9 to bind RNA.
- A preliminary pipeline was built, docking between protein and ssRNA structures is implemented successfully:
- Docking is implemented between the rigid fixed protein structure and 1 ssRNA structure, with a manually chosen coordinates;
- Active residue(s) of a protein can be selected;
- The score is calculated for an initial (before the docking) and resulting (after the docking) pose.
- In the web interface were implemented:
-
Sliders, which allow choosing initial RNA position;
-
Space to view the score of the initial pose and the resulting pose;
-
Space to view and manipulate pdb structures, including initial pose and resulting pose;
-
Space to select active protein residues;
-
Visualization of the status graph, provided by Seamless framework.
The listed points allow performing actions in collaboration with the remote participants.
- Scientific:
- Continued study of conserved residues;
- Search for examples of mutations that increase the binding between protein and ssRNA: currently, most of the mutations we found decrease the binding affinity.
- Pipeline:
- with the help of the Cloudless, add the possibility to launch independent instances of the pipeline and share these instances with remote users.
- Web interface:
- add a possibility to upload the pdb files to the pipeline from the local machine through the web-interface;
- make an interface aesthetically pleasing
At the end of the coding session, assign the role for the next coding session
-
Will make a screenshot of a status graph after each session
-
Will update README.md and is responsible for task list
- Keeps an eye on questions in the chat
This month we still Google Docs, use VMD/Pymol session
On longer term => Markdown on Git, use our workflow web interface for visualization
In the near/medium term: a permanent link where the workflow (web form + status graph) is published for everyone