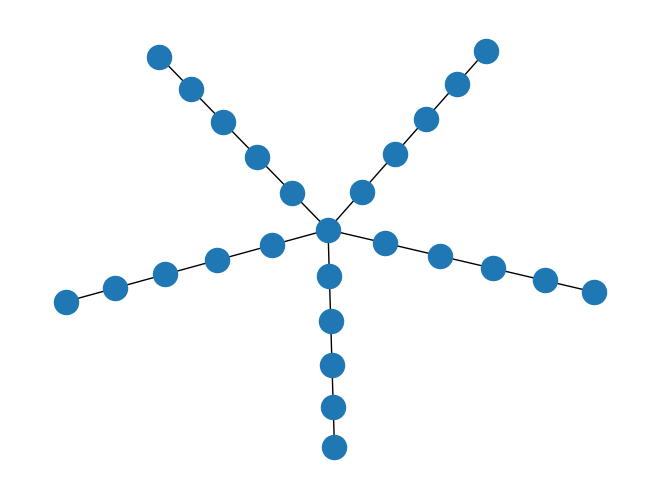
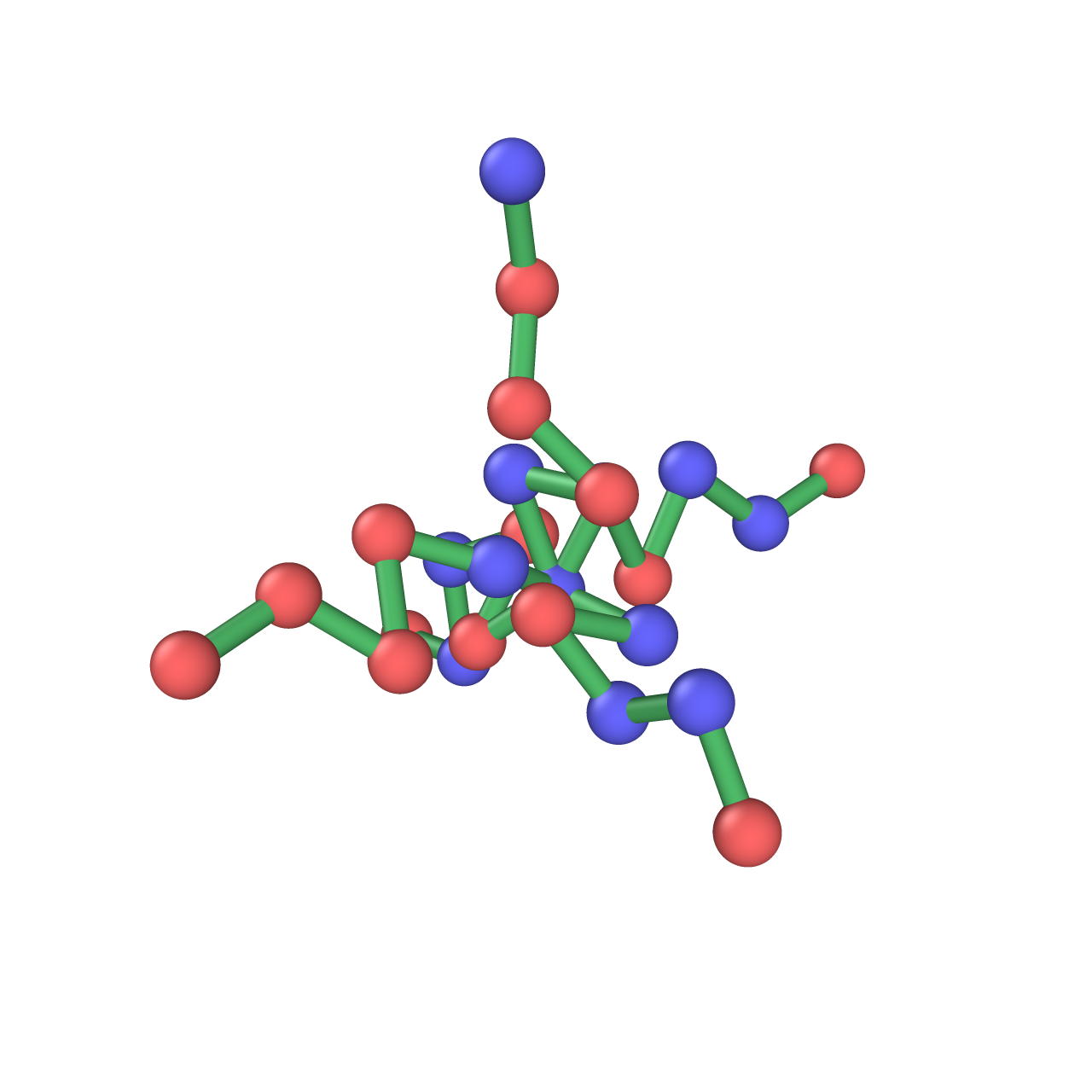
This program enables the generation of LAMMPS input data based on networkx graphs.
Installation with pip:
git clone git@github.com:sjiang87/graph2lammps.git
cd graph2lammps
pip install -e .import numpy as np
import networkx as nx
from graph2lammps.convert import gen_pos, write_pdb, write_lammps-
Create a polymer graph, e.g., a star polymer:
def gen_star(num_arm, len_branch): G = nx.Graph() G.add_node(0) for i in range(1, num_arm + 1): arm = nx.path_graph(len_branch) arm = nx.relabel_nodes(arm, {j: i*len_branch + j for j in range(len_branch)}) G = nx.union(G, arm) G.add_edge(0, i*len_branch) return G G = gen_star(num_arm=5, len_branch=5) mapping = {old_label: new_label for new_label, old_label in enumerate(sorted(G.nodes()))} G = nx.relabel_nodes(G, mapping)
-
Assign properties to each node or edge. This can be done after graph creation or directly in the custom 'gen_star' function.
for node in G.nodes(): G.nodes[node]['type'] = random.randint(1, 2) # bead type 1 or 2 G.nodes[node]['name'] = string.ascii_uppercase[G.nodes[node]['type'] ] # some random node names for edge in G.edges(): G.edges[edge]['type'] = random.randint(1, 2) # bond type 1 or 2 G.graph['mass'] = {1: 1, 2:2} # graph-level metadata, e.g., bead mass, charge
-
Create node position matrix:
pos = gen_pos(G, rmin=0.5, bdist=1.0, bmin=0.9, bmax=1.1, niter=1000)
rmin : float Minimum separation distance between polymer beads. bdist : float Ideal bond distance between connected polymer beads. bmin : float Minimum bond length. bmax : float Maximum bond length. niter : int Number of iterations for position generation. -
Write LAMMPS data file.
write_lammps(G, pos, fname='your/dir/sys.data')
The above-mentioned procedure can be completed by:
from graph2lammps import graph2lammps
G = gen_star(num_arm=5, len_branch=5)
graph2lammps(G, rmin=0.5, bdist=1.0, bmin=0.9, bmax=1.1, niter=1000, fname='your/dir/sys.data)- Currently supports bead/atom and bond types. Support for angles, dihedrals, and other types will be added.
- An executable command will be added to convert a pickled file containing multiple graphs to multiple LAMMPS data files.
If you need to add additional functionality, please contact sj0161@princeton.edu or submit a pull request.