Ye Zheng*, Siqi Shen* and Sündüz Keleş. Normalization and De-noising of single-cell Hi-C Data with BandNorm and 3DVI. bioRxiv (2021). * contribute equally.
Siqi Shen, Ye Zheng* and Sündüz Keleş*, scGAD: single-cell gene associating domain scores for exploratory analysis of scHi-C data. bioRxiv (2021). * corresponding authors.
The advent of single-cell sequencing technologies in profiling 3D genome organization led to development of single-cell high-throughput chromatin conformation (scHi-C) assays. Data from these assays enhance our ability to study dynamic chromatin architecture and the impact of spatial genome interactions on cell regulation at an unprecedented resolution. At the individual cell resolution, heterogeneity driven by the stochastic nature of chromatin fiber, various nuclear processes, and unwanted variation due to sequencing depths and batch effects poses major analytical challenges for inferring single cell-level 3D genome organizations.
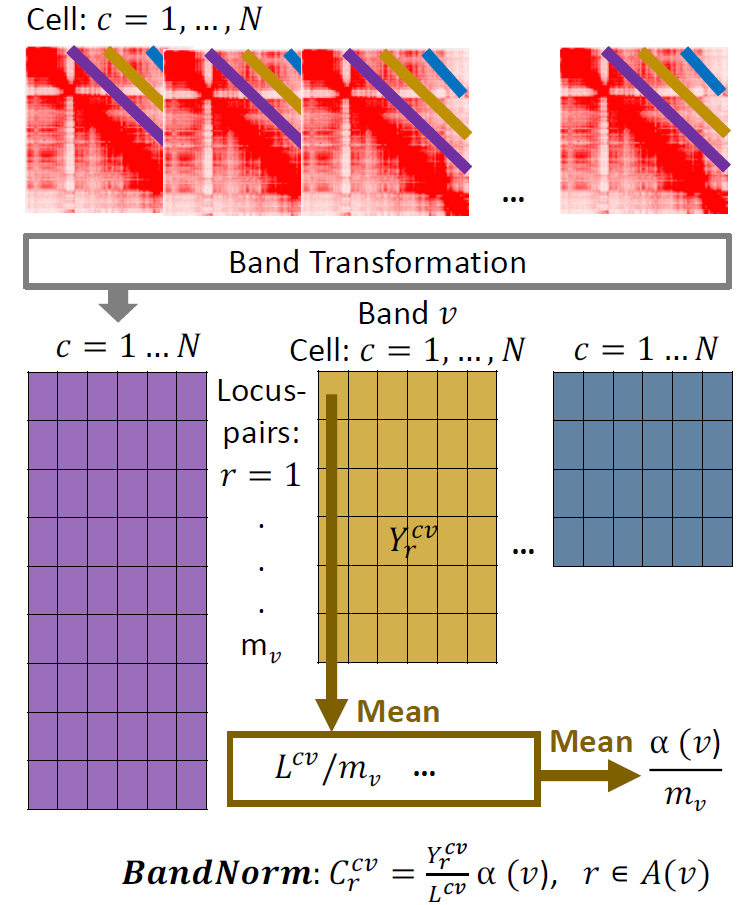
To explicitly capture chromatin conformation features and distinguish cells based on their 3D genome organizations, we develop a simple and fast band normalization approach, BandNorm, as well as a deep generative modeling framework, 3DVI, for more structured modeling of scHi-C data. BandNorm first removes genomic distance bias within a cell, and sequencing depth normalizes between cells. Consequently, BandNorm adds back a common band-dependent contact decay profile for the contact matrices across cells. The former step is achieved by dividing the interaction frequencies of each band within a cell with the cell’s band mean. The latter step is implemented by multiplying each scaled band by the average band mean across cells.
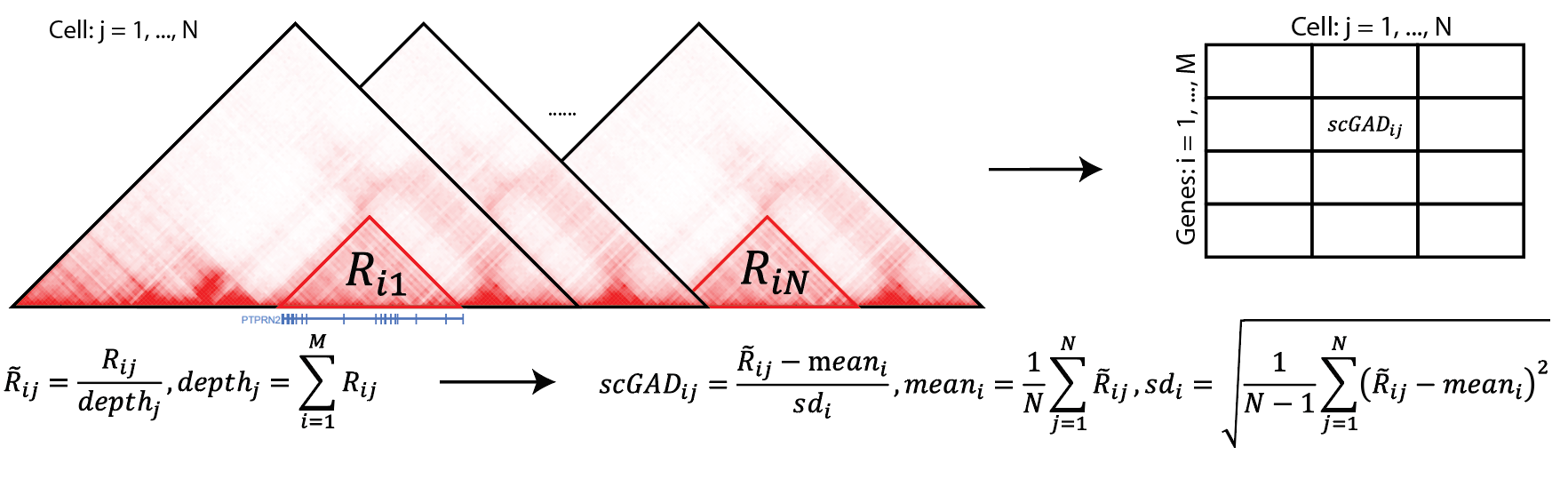
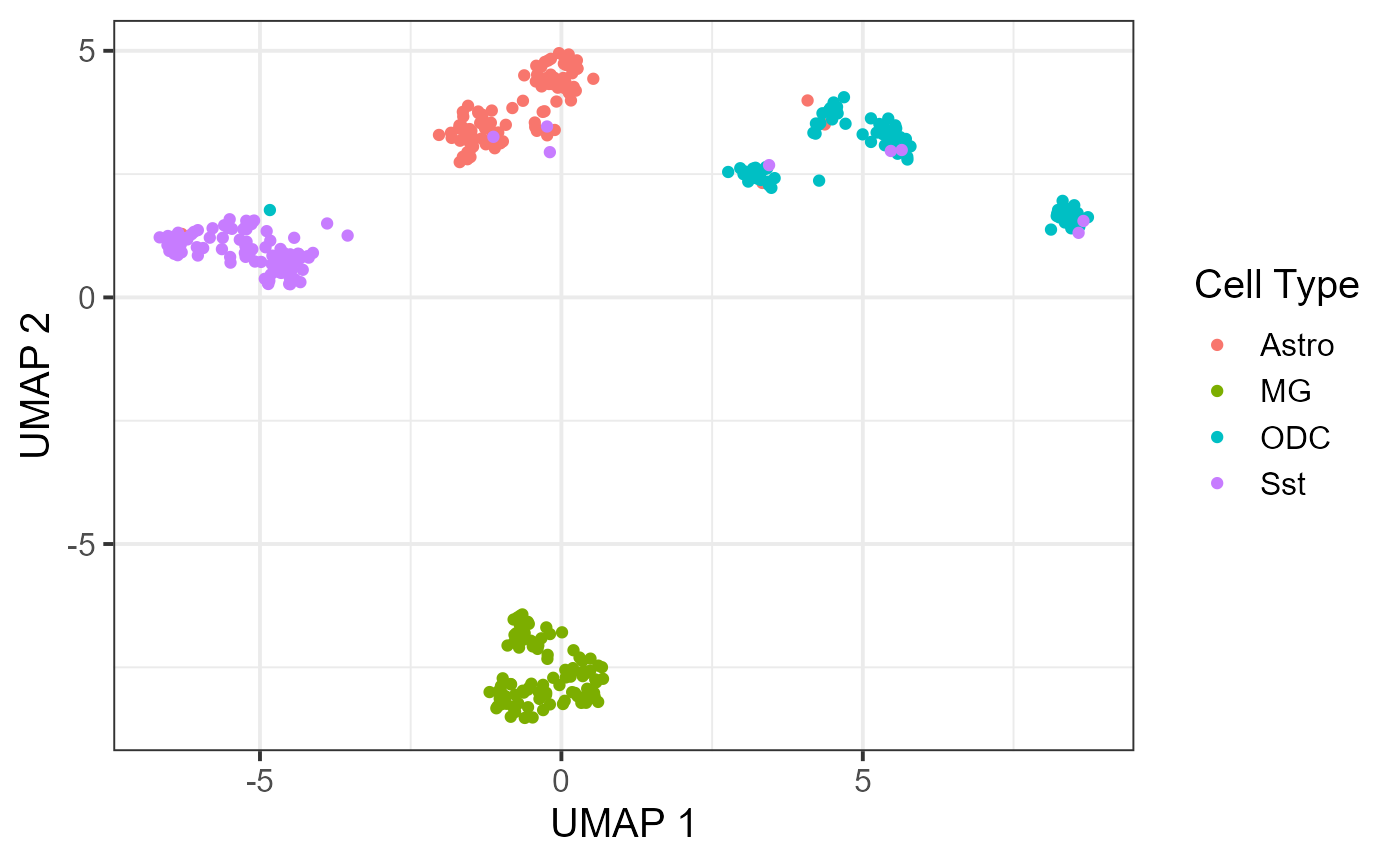
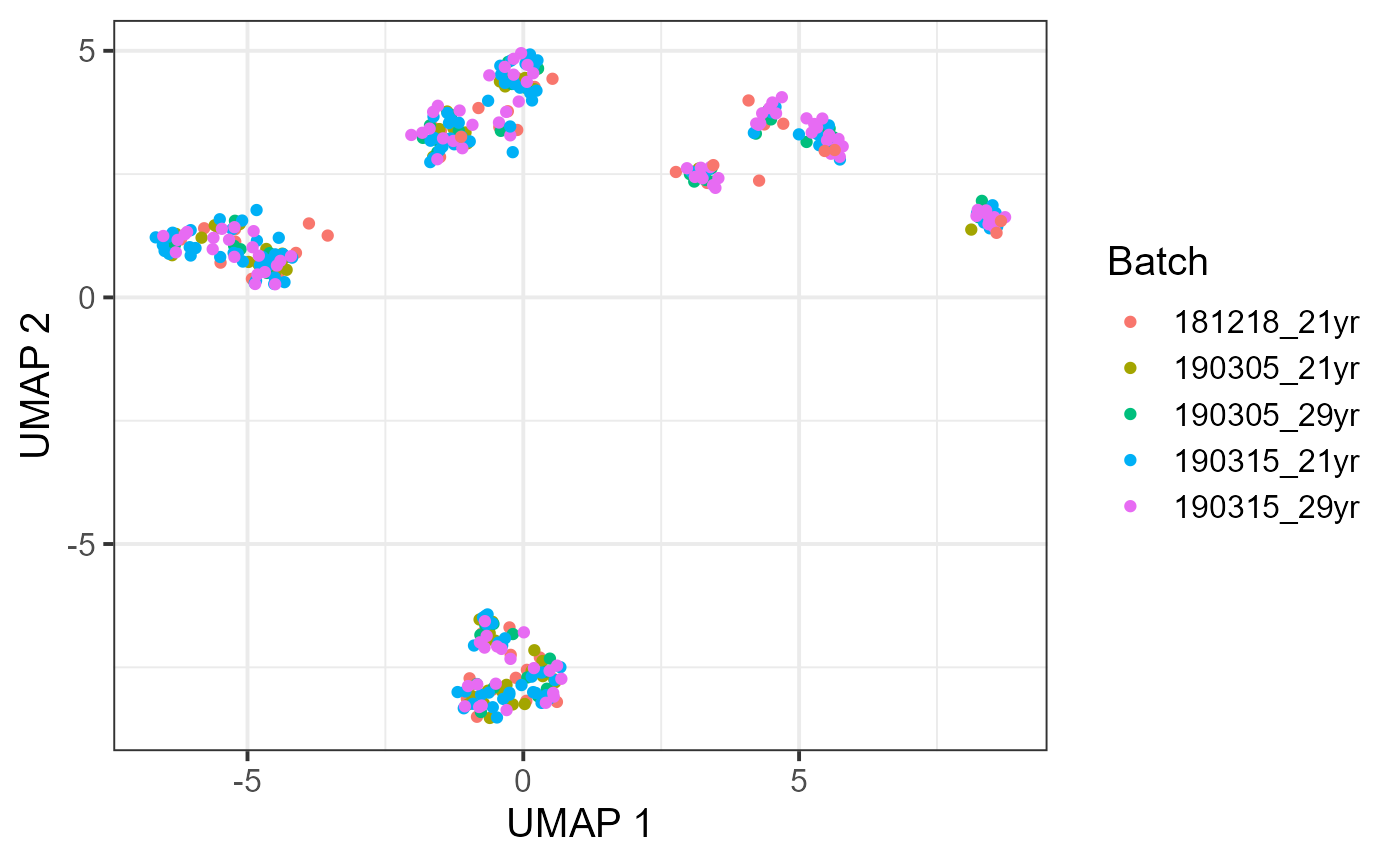
Recent advancements in single-cell technologies enabled the profiling of 3D genome structures in a single-cell fashion. Quantitative tools are needed to fully leverage the unprecedented resolution of single-cell high-throughput chromatin conformation (scHi-C) data and integrate it with other single-cell data modalities. We present single-cell gene associating domain (scGAD) scores as a dimension reduction and exploratory analysis tool for scHi-C data. scGAD enables summarization at the gene level while accounting for inherent gene-level genomic biases. Low-dimensional projections with scGAD capture clustering of cells based on their 3D structures. scGAD enables identifying genes with significant chromatin interactions within and between cell types. We further show that scGAD facilitates the integration of scHi-C data with other single-cell data modalities by enabling its projection onto reference low-dimensional embeddings such as scRNA-seq.
There are seven functions, download_schic, bandnorm, bandnorm_juicer, create_embedding, and plot_embedding, scGAD and runProjection.
download_schic downloads one of the currently available single-cell Hi-C data cleaned by us, bandnorm takes in sparse matrices and normalize them using BandNorm method, and bandnorm_juicer allows using .hic data to perform BandNorm, create_embedding summarizes the data into a PCA embedding in preparation for clustering and lower dimension embedding, plot_embedding calculates UMAP or tSNE embedding from the PCA obtained from create_embedding, and plots the resulting embedding. scGAD obtains scGAD score from single-cell Hi-C data, and runProjection projects scGAD score on other single-cell modalities.
(See vignette (browseVignettes("BandNorm")) or visit BandNorm website for more detail.)
If necessary, install the dependencies:
install.packages(c('ggplot2', 'dplyr', 'data.table', 'Rtsne', 'umap'))
if (!requireNamespace("devtools", quietly=TRUE))
install.packages("devtools")
library(devtools)
install_github("immunogenomics/harmony")
BandNorm can be installed from Github (User can change build_vignettes to TRUE to locally compile the vignettes. Note that the vignettes are also available online at BandNorm tutorial and scGAD tutorial (BandNorm Method for single-cell Hi-C • BandNorm (sshen82.github.io)):
devtools::install_github('sshen82/BandNorm', build_vignettes = FALSE)
library(BandNorm)
Also, if the user doesn't have default R path writing permission, it is possible to install the package to the personal library:
withr::with_libpaths("personalLibraryPath", devtools::install_github("sshen82/BandNorm"))
Below are the outputs of the code in the vignette.
For more detail, please visit BandNorm website.