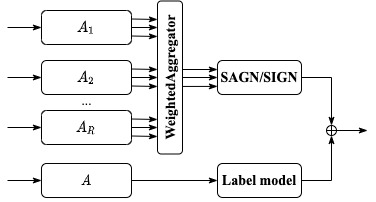
This is an implementation of a simple combination of SAGN+SLE and NARS.
Since DGLKE does not support the latest DGL, there exist different requirements of python package for preprocessing and training. We suggest creating two associated conda environments.
The main python packages for preprocessing:
ogb==1.3.1
pytorch==1.8.1
dgl==0.4.3.post2
dglke==0.1
The main python packages for training:
ogb==1.3.1
pytorch==1.8.1
dgl==0.6.1
We move graph operations for convolution and label propagation to the CPU, which may require extra memory (>25Gb).
We simply combine NARS and SAGN+SLE.
NARS can be viewed as a heterogeneous version of SIGN.
To proceed heterographs, NARS randomly samples relations subsets to generate a "multi-relation" view for nodes. For example, in the MAG (ogbn-mag) dataset, there are four types of relations (cites, writes, affliated_with, has_topic). We can randomly select relations to generate different subsets of relations (["cites", "writes"], ["writes"], ["cites", "affiliated_with", "has_topic"] and so on). Note that the sampled subsets should have access to target nodes (paper nodes). Thus, there are 11 valid subsets in total. To reduce memory footprints, we can generate fewer subsets in each run. Each subset can be viewed as a subgraph (see K+1 node feature matrices of different aggregation order (hop). Suppose that we generate R subsets. Then we have (K+1)*R feature matrices. Although the 0-hop (raw) feature matrix appears multiple times.
To apply SIGN based on these matrix while preserving considerable complexity, NARS introduces a 1D convolution. For each order (hop), it simply sums matrices from different subsets with learnable weights. These weights are stored in a (R,(K+1),D) tensor. This operation is implemented as a WeightedAggregator module. It outputs K+1 feature matrices which can be directly fed into an SIGN module.
In addition, the NARS paper also points out that pretrained methods for graph embedding such as TransE and Metapath2Vec can bring significant improvements for scalable methods on heterographs. In this repository, we use TransE embeddings as paddings for non-attributed nodes.
SLE is a training framework designed for scalable or sampling-free methods such as SGC, SIGN, SAGN and MLP. It combines self-training and label propagation.
For a base model, we combine it with a simple label model whose input is smoothed label embedding. The label model is designed as a simple MLP. We sum up the outputs of the base model and label model.
At the first stage, we train a model following standard processing. From the second stage, we train a new model with an enhanced train set. We filter confident nodes with previously predicted probabilities. Then we select the union of raw train set and confident nodes set as the enhanced train set. The extra nodes in the enhanced train set use their pseudo hard labels as target labels. After several stages, we obtain the final model.
Besides NARS's weighted aggregation, we also propose a naive HSAGN which directly takes all feature matrices as input. This model is used in the OGB-LSC MAG240M challenge with 72.70% validation accuracy without using validation as train data or ensemble tricks.
SAGN is a more adaptive version of SAGN. In short, SAGN uses a learnable attention mechanism among hops to replace the concatenation operation in SIGN. It is intuitive to use SAGN instead of SIGN in NARS to obtain better expressiveness, which is denoted by NARS_SAGN. In addition, to unify names, the original NARS is denoted by NARS_SIGN.
Then we can use NARS_SAGN as the base model in SLE. In each run, we firstly perform NARS's subset sampling (or we can use fixed subsets). These subsets are used across different stages.
In total, first we design a combined model with NARS_SAGN and node label model (MLP) as the base model. Note that we use the whole graph to perform label propagation.
Then we perform preprocessing (additional dataset processing, TransE pretraining, subset sampling if needed) in each run.
At each stage, we perform neighbor averaging and label propagation. At the first stage, we train the first model with the raw train set. From the second stage, we enhance the training set and train a new model. Finally, we obtain the best model.
You can generate all the necessary pretrained embeddings using scripts from NARS.
For example, suppose that we want to train models on MAG.
Firstly, we train transE following instructions in NARS/graph_embed.
Then we can train our models with main.py.
For OAG datasets, you should download and preprocess them by following the instructions from NARS.
These arguments should be modified by your own paths:
--root: The root directory for ogb datasets.
--embedding-path: The root directory of TransE embedding. If your embedding .pt file is saved under NARS/TransE_mag, then you should set it to NARS.
--example-subsets-path: The directory of example relations subsets (only used when `fixed-subsets` is True).
Some key hyperparameters:
--model: "hsagn" is a naive heterogeneous version of SAGN.
"nars_sagn" replaces the SIGN component with SAGN in NARS.
"nars_sign" is the original NARS.
--K: The maximum neighbor averaging hop.
--label-K: The maximum hop for label propagation.
--sample-size: The number of sampled relation subsets.
--fixed-subsets: Whether to use example subsets from NARS/sample_relation_subsets/examples.
We provide example scripts for experiments on ACM, MAG, OAG_venue and OAG_L1 in directory scripts. The full hyperparameter settings are included in these scripts. For now, these experiments have not been fully conducted. NARS_SAGN has worse results at the first stage but better results after two stages. The label information seems less important for ACM.
Note: To reproduce the reported best results on MAG, please execute the script mag_sagn_use_labels_fixed.sh.
| Relation size | Model | Dataset | Test acc | Val acc |
|---|---|---|---|---|
| fixed 8 | NARS_SAGN+0-SLE | MAG | 0.5232±0.0025 | 0.5412±0.0015 |
| fixed 8 | NARS_SAGN+1-SLE | MAG | 0.5395±0.0014 | 0.5552±0.0016 |
| fixed 8 | NARS_SAGN+2-SLE | MAG | 0.5440±0.0015 | 0.5591±0.0017 |
| 8 | NARS_SIGN+0-SE | MAG | 0.5173±0.0032 | 0.5323±0.0050 |
| 8 | NARS_SIGN+1-SE | MAG | 0.5230±0.0067 | 0.5319±0.0076 |
| 8 | NARS_SIGN+2-SE | MAG | 0.5218±0.0087 | 0.5312±0.0088 |
| 8 | NARS_SAGN+0-SE | MAG | 0.5052±0.0033 | 0.5205±0.0037 |
| 8 | NARS_SAGN+1-SE | MAG | 0.5204±0.0040 | 0.5311±0.0054 |
| 8 | NARS_SAGN+2-SE | MAG | 0.5224±0.0050 | 0.5327±0.0058 |
| 8 | NARS_SIGN+0-SLE | MAG | 0.5232±0.0024 | 0.5411±0.0032 |
| 8 | NARS_SIGN+1-SLE | MAG | 0.5347±0.0049 | 0.5508±0.0052 |
| 8 | NARS_SIGN+2-SLE | MAG | 0.5382±0.0051 | 0.5546±0.0046 |
| 8 | NARS_SAGN+0-SLE | MAG | 0.5207±0.0039 | 0.5400±0.0028 |
| 8 | NARS_SAGN+1-SLE | MAG | 0.5370±0.0038 | 0.5544±0.0037 |
| 8 | NARS_SAGN+2-SLE | MAG | 0.5408±0.0042 | 0.5577±0.0034 |
| Relation size | Model | Dataset | Test acc | Val acc |
|---|---|---|---|---|
| 2 | NARS_SIGN+0-SE | ACM | 0.9230±0.0062 | 0.9289±0.0106 |
| 2 | NARS_SIGN+1-SE | ACM | 0.9341±0.0031 | 0.9476±0.0090 |
| 2 | NARS_SIGN+2-SE | ACM | 0.9391±0.0050 | 0.9392±0.0136 |
| 2 | NARS_SAGN+0-SE | ACM | 0.9166±0.0054 | 0.9279±0.0092 |
| 2 | NARS_SAGN+1-SE | ACM | 0.9358±0.0062 | 0.9481±0.0098 |
| 2 | NARS_SAGN+2-SE | ACM | 0.9415±0.0061 | 0.9414±0.0144 |
| 2 | NARS_SIGN+0-SLE | ACM | 0.9216±0.0043 | 0.9279±0.0089 |
| 2 | NARS_SIGN+1-SLE | ACM | 0.9324±0.0062 | 0.9439±0.0060 |
| 2 | NARS_SIGN+2-SLE | ACM | 0.9387±0.0040 | 0.9392±0.0152 |
| 2 | NARS_SAGN+0-SLE | ACM | 0.9173±0.0044 | 0.9279±0.0078 |
| 2 | NARS_SAGN+1-SLE | ACM | 0.9324±0.0051 | 0.9471±0.0096 |
| 2 | NARS_SAGN+2-SLE | ACM | 0.9394±0.0090 | 0.9419±0.0135 |
| Relation size | Model | Dataset | Test MRR | Val MRR | Test NDCG | Val NDCG |
|---|---|---|---|---|---|---|
| 8 | NARS_SIGN+0-SE | OAG_venue | 0.3612±0.0052 | 0.4293±0.0046 | 0.5380±0.0047 | 0.6051±0.0039 |
| 8 | NARS_SIGN+1-SE | OAG_venue | 0.3736±0.0053 | 0.4362±0.0040 | 0.5499±0.0046 | 0.6107±0.0034 |
| 8 | NARS_SIGN+2-SE | OAG_venue | 0.3745±0.0050 | 0.4358±0.0041 | 0.5498±0.0042 | 0.6093±0.0032 |
| 8 | NARS_SAGN+0-SE | OAG_venue | 0.3517±0.0047 | 0.4179±0.0033 | 0.5271±0.0042 | 0.5940±0.0030 |
| 8 | NARS_SAGN+1-SE | OAG_venue | 0.3633±0.0051 | 0.4233±0.0037 | 0.5379±0.0047 | 0.5985±0.0034 |
| 8 | NARS_SAGN+2-SE | OAG_venue | 0.3647±0.0064 | 0.4225±0.0034 | 0.5391±0.0061 | 0.5972±0.0030 |
| 8 | NARS_SIGN+0-SLE | OAG_venue | 0.3622±0.0049 | 0.4470±0.0045 | 0.5354±0.0046 | 0.6167±0.0041 |
| 8 | NARS_SIGN+1-SLE | OAG_venue | 0.3808±0.0058 | 0.4588±0.0045 | 0.5537±0.0056 | 0.6274±0.0038 |
| 8 | NARS_SIGN+2-SLE | OAG_venue | 0.3834±0.0063 | 0.4589±0.0049 | 0.5549±0.0061 | 0.6262±0.0041 |
| 8 | NARS_SAGN+0-SLE | OAG_venue | 0.3536±0.0043 | 0.4397±0.0040 | 0.5258±0.0039 | 0.6096±0.0034 |
| 8 | NARS_SAGN+1-SLE | OAG_venue | 0.3716±0.0047 | 0.4517±0.0036 | 0.5434±0.0043 | 0.6206±0.0032 |
| 8 | NARS_SAGN+2-SLE | OAG_venue | 0.3742±0.0063 | 0.4527±0.0040 | 0.5451±0.0058 | 0.6209±0.0037 |