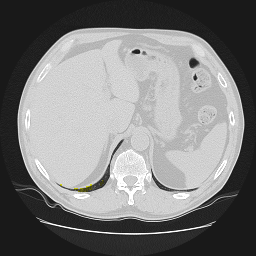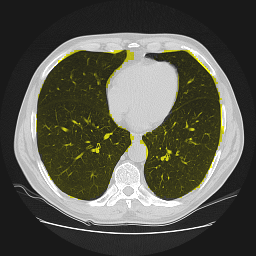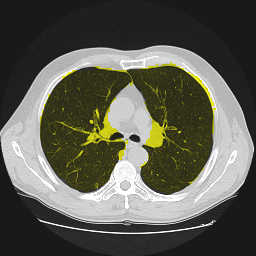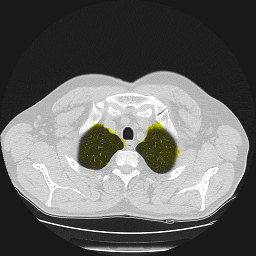
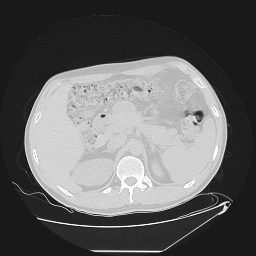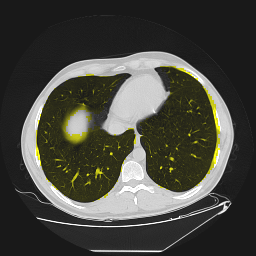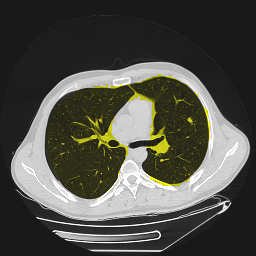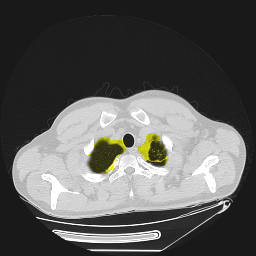
Segmentation of lungs in 3D Computed Tomography (CT) scans via non-rigid image registration scheme [1].
Short description of the current implementation can be found in [2] (please consider citing if using this implementation).
Running the scripts requires
- Python 3.6+ with the following packages installed (all installable via pip):
numpy
scikit-image
scipy
pandas
nibabel
imageio
- Elastix tool installed in your
PATH. The tool can be downloaded from the website (http://elastix.isi.uu.nl/) or installed on Unix systems via:
sudo apt install elastix
NOTE: the Elastix official website seems to be not working, therefore you can try to download Elastix for Windows here.
- Download the CT_RegSegm_data.tar (289 MB)
archive and extract its content (
resized_dataandtest_data) into the repository root directory.
The scripts can be run directly from Python (see go_c_regsegm.py) or from command line using run_regsegm.py script. Examples:
python3 run_regsegm.py test_data/test_image.nii.gz
python3 run_regsegm.py test_data/dir_with_images
If succeeded, the *_regsegm_py.nii.gz files containing the lung masks should appear next to the original files.
Result examples:
[1] Sluimer I, Prokop M, van Ginneken B. Toward automated segmentation of the pathological lung in CT. IEEE Trans Med Imaging 2005;24(8):1025–1038. (URL: https://www.ncbi.nlm.nih.gov/pubmed/16092334)
[2] Liauchuk, V., Kovalev, V.: ImageCLEF 2017: Supervoxels and co-occurrence for tuberculosis CT image classification. In: CLEF2017 Working Notes. CEUR Workshop Proceedings, Dublin, Ireland, CEUR-WS.org http://ceur-ws.org (September 11-14 2017) (URL: http://ceur-ws.org/Vol-1866/paper_146.pdf).