This external app displays compound ids and their properties with highlighted rows based on a date range.
Specifically, it ingests query parameters, -TABLENAME- passed from DM UI to
pre-populate a textarea with compound ids that are to be viewed for structure activity relationship analysis. The -TABLENAME- mask will direct the backend to select the COMPOUND_ID column from the corresponding query table.
The python fastapi backend is used to generate the SQL and run it against the connected Oracle database. The application was deployed using
Jenkins CI/CD pipline. The Jenkinsfile can be inspected to understand what is performed. Basically, the docker image is built, then pushed to ECR, then deployed using helm. The app lives in the main apps namespace within helm
as well as kubectl. Run kubectl get all -n apps -l app=sar-view to get the kubernetes resources. Currently, the app can be visited by navigating to: http://sar-view.kinnate.
The backend architecture uses Celery workers that are deployed manually using kubectl commands. The deployment yaml looks like:
apiVersion: apps/v1
kind: Deployment
metadata:
name: sar-view-${DEV}celery-worker
namespace: apps
labels:
app: sar-view-${DEV}celery-worker
spec:
replicas: $REPLICA_COUNT
selector:
matchLabels:
app: sar-view-${DEV}celery-worker
template:
metadata:
labels:
app: sar-view-${DEV}celery-worker
spec:
containers:
- name: celery-worker
image: ${AWSID}.dkr.ecr.us-west-2.amazonaws.com/sar-view-${DEV}backend:latest
imagePullPolicy: Always
env:
- name: REDIS_HOST
value: '$REDIS_HOST'
- name: REDIS_PASSWD
valueFrom:
secretKeyRef:
name: redis-secret
key: redis_password
command: ['celery']
args: ['-A', 'app.celery', 'worker', '--loglevel=info']
resources:
limits:
cpu: 400m
requests:
cpu: 200m
Simply pass the values using REPLICA_COUNT=2 DEV=test- REDIS_HOST=redis-svc-dev-master.redis envsubst < deploy.yaml | k apply -f - to deploy it to the kubernetes cluster. Afterwards, deploy an HPA for better pod efficiency. Since the backend executes a long running process, it was decided to use HPA to keep up with the load.
apiVersion: autoscaling/v1
kind: HorizontalPodAutoscaler
metadata:
name: sar-view-${DEV}celery-worker
namespace: apps
spec:
scaleTargetRef:
apiVersion: apps/v1
kind: Deployment
name: sar-view-${DEV}celery-worker
minReplicas: $REPLICA_COUNT
maxReplicas: 10
targetCPUUtilizationPercentage: 50
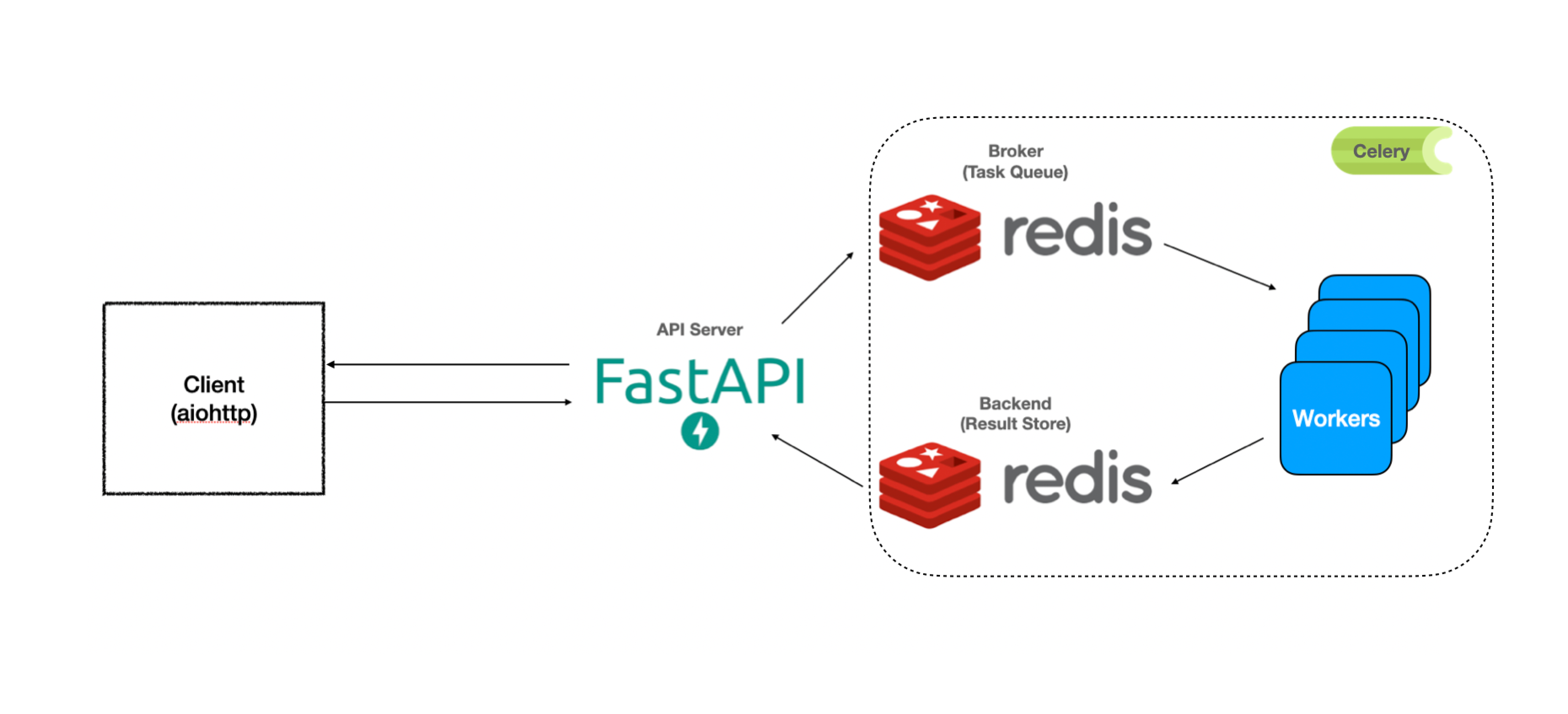
The following diagram shows the architecture of the backend. fastapi initiates a
background task which then awaits the response from the redis cluster. The redis cluster is updated in parallel by the celery worker that off-loads the background task.
Once jenkins deploys the app and the pods and services are running the next step
is to expose the application through a kubernetes nginx ingress. Just as in any other app, one must add a new ingress rule to the
ingress yaml file. This can be obtained by running: kubectl get ingress -o yaml and applyling it locally, or editing the yaml on the fly (expert user)
like so: kubectl edit ingress -n apps. The ingress rule block should resemble
something like:
- host: $APP_NAME.kinnate
http:
paths:
- path: /
pathType: Prefix
backend:
service:
name: $APP_NAME-svc
port:
name: http
Then ensure this A-record is added to Route 53 with the appropriate LoadBalancer
name as the Route traffic to value.
The biochemical geomean SQL statement is extracted from http://sql-ds.kinnate
service and formatted properly for the SAR view. If changes were made to the SQL
source on the Dotmatics side, use this endpoint to update the SQL:
http://sar-view-backend.kinnate/v1/update_sql_ds. Restarting the pods will also achieve the same result since it will retrieve the SQL on start-up. Test the sql-datasource service after making changes to the SQL on the DM side.
An example payload would be:
{"biochemical": {"id": 912,"app_type": "geomean_sar"}}