The MuSyc app in its current state only predicts using the MuSyc model. Future developments will integrate the Loewe and HSA models. The backend is a python fastapi app that borrowed core code from https://github.com/djwooten/synergy.
The current fit algorithm leveraged in the MuSyC portal is described in the Methods section of Wooten et al. 2020. The authors use a Monte Carlo algorithm as suggested by Motulsky and Christopoulos (Chapter 17, pg 104) for estimating asymmetric 95% confidence intervals of each parameter. Briefly, this is done by fitting all the data using standard non-linear least squares regression (TFR option in SciPy's curve_fit). Based on this optimal fit, noise is added to every data point proportional to the root mean square error of the optimal fit. The new "noise-added" data is then fit again to generate a new parameter set. This process is run 100 times and the 95% confidence intervals for all parameters are calculated from the ensemble.
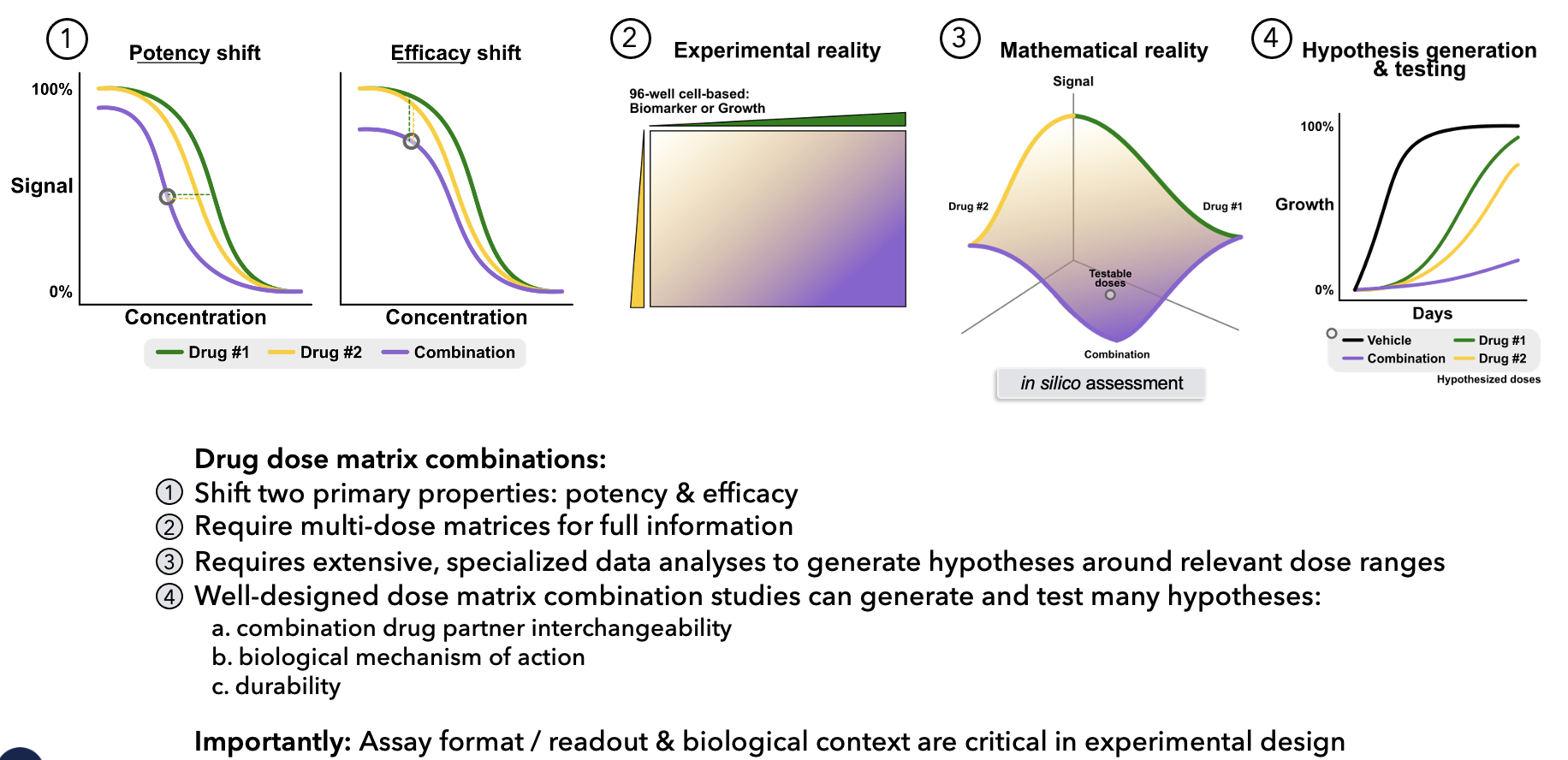
- U - Percent of unaffected population
- A1, A2 - Percent of affected by drug 1 and drug 2, respectively
- A1,2 - Percent of affected by both drug 1 and drug 2
- d1, d2 - Drug concentrations for drug pair
- Ed - Measured effect at (d1, d2)
- C1, C2 - The concentration of drug required to achieve 50% of the
- maximal effect (i.e., EC50)
- h1, h2 - Hill coefficients for dose-response curves of drug 1 and 2 in
- isolation
- E0 - The basal effect Ed (d1 = d2 = 0)
- E1, E2 - Maximal efficacy of drugs 1 and 2 in isolation
- E3 - Maximal efficacy of the combination of drugs 1 and 2
- β - Percent increase (or decrease) in max effect with both drugs over the most efficacious single drug (β = (min(E1,E2)-E3)/E0-min(E1,E2))
- α12 - Fold change in the potency (C2) of [d2] induced by drug 1
- α21 -Fold change in the potency (C1) of [d1] induced by drug 2
- γ12 - Fold change in the cooperativity (h2) of [d2] induced by drug 1
- γ21 - Fold change in the cooperativity (h1) of [d1] induced by drug 2
- https://www.nature.com/articles/s41467-021-24789-z
- https://academic.oup.com/bioinformatics/article/37/10/1473/5909985?login=false
- https://github.com/djwooten/natcomms-musyc2021
- https://github.com/djwooten/synergy
This app allows users to drag n' drop .csv files with exactly 5 columns: (1)
drug1.conc, drug2.conc, drug1.name, drug2.name and effect, which will
generate a Score table of the beta, alpha12, alpha21, gamma12, and
gamma21 coefficients. There is code to generate a 2D heatmap as well as a 3D
surface plot, however that has not been entirely implemented for production,
only for development and testing.
Run npm run start in frontend directory. Run python main.py in backend directory. Ensure the python environemnt has all the libraries from requirements.txt. Access localhost:3000 which will point to the CsvUpload component. Users can drag n' drop .csv files to generate the Score table. There is a Dockerfile in the postgres directory that would need to be built in order to use the public dataset for testing. Once the image is available and the container is named postgres, the python backend should be able to access it. If tmux is installed, one can use the startup.sh script to automatically start the application in its entirety.
ReactJS frontend and fastapi python backend. Postgres is a dependency for testing
of public sample datasets extracted from: http://drugcombdb.denglab.org/. Source file: drugcombs_response.csv
Browse to http://localhost:3000/test?compound_id=${FT_NUMBER}
where FT_NUMBER is any valid compound id that is stored in database. Currently as of writing this documentation, it is a fixed list that is saved in the backend application, named ft_nbrs.py