corona19
R package for import datasets of Coronavirus disease 2019 in South Korea from Data-Science-for-COVID-19.
Installation
# install.packages("remotes")
remotes::install_github("youngwoos/corona19")Example
library(corona19)
# time
time <- getdata("time")
time
#> # A tibble: 57 x 7
#> date time test negative confirmed released deceased
#> <date> <int> <int> <int> <int> <int> <int>
#> 1 2020-03-16 0 274504 251297 8236 1137 75
#> 2 2020-03-15 0 268212 243778 8126 834 75
#> 3 2020-03-14 0 261335 235615 8086 714 72
#> 4 2020-03-13 0 248647 222728 7979 510 67
#> 5 2020-03-12 0 234998 209402 7869 333 66
#> 6 2020-03-11 0 222395 196100 7755 288 60
#> 7 2020-03-10 0 210144 184179 7513 247 54
#> 8 2020-03-09 0 196618 171778 7382 166 51
#> 9 2020-03-08 0 188518 162008 7134 130 50
#> 10 2020-03-07 0 178189 151802 6767 118 44
#> # ... with 47 more rows
library(ggplot2)
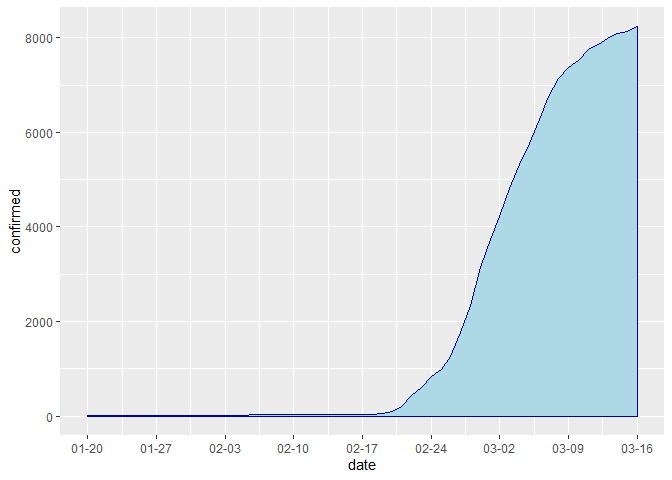
ggplot(data = time, aes(x = date, y = confirmed)) +
geom_area(color="darkblue", fill="lightblue") +
scale_x_date(date_breaks = "weeks" , date_labels = "%m-%d")# patient
patient <- getdata("patient")
patient
#> # A tibble: 2,119 x 18
#> patient_id global_num sex birth_year age country province city disease
#> <dbl> <int> <chr> <int> <chr> <chr> <chr> <chr> <lgl>
#> 1 1000000001 2 male 1964 50s Korea Seoul Gang~ NA
#> 2 1000000002 5 male 1987 30s Korea Seoul Jung~ NA
#> 3 1000000003 6 male 1964 50s Korea Seoul Jong~ NA
#> 4 1000000004 7 male 1991 20s Korea Seoul Mapo~ NA
#> 5 1000000005 9 fema~ 1992 20s Korea Seoul Seon~ NA
#> 6 1000000006 10 fema~ 1966 50s Korea Seoul Jong~ NA
#> 7 1000000007 11 male 1995 20s Korea Seoul Jong~ NA
#> 8 1000000008 13 male 1992 20s Korea Seoul etc NA
#> 9 1000000009 19 male 1983 30s Korea Seoul Song~ NA
#> 10 1000000010 21 fema~ 1960 60s Korea Seoul Seon~ NA
#> # ... with 2,109 more rows, and 9 more variables: infection_case <chr>,
#> # infection_order <int>, infected_by <chr>, contact_number <int>,
#> # symptom_onset_date <date>, confirmed_date <date>, released_date <date>,
#> # deceased_date <date>, state <chr>
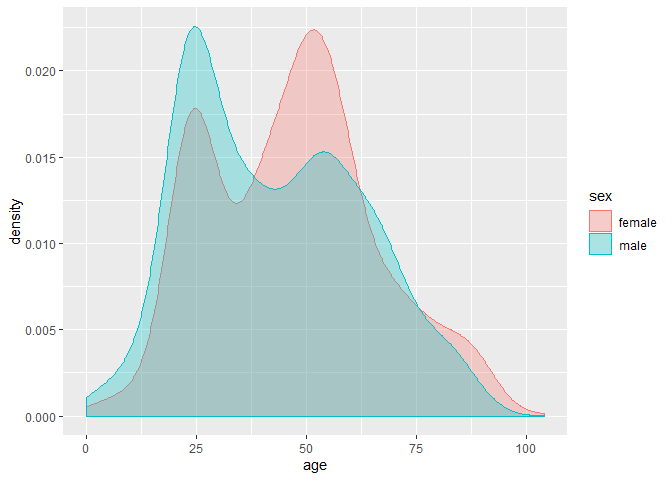
ggplot(data = patient, aes(x = 2020-birth_year, fill = sex, colour = sex)) +
geom_density(alpha = 0.3) +
xlab("age")# route
route <- getdata("route")
library(dplyr)
cnt_province <- route %>%
count(province, sort = T) %>%
head(10)
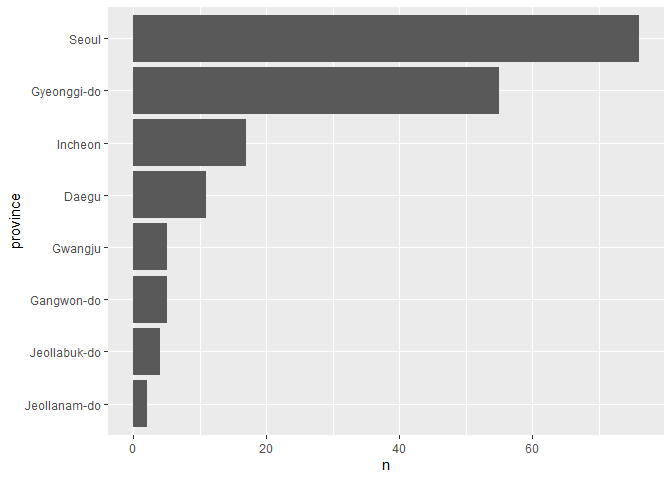
cnt_province
#> # A tibble: 8 x 2
#> province n
#> <chr> <int>
#> 1 Seoul 76
#> 2 Gyeonggi-do 55
#> 3 Incheon 17
#> 4 Daegu 11
#> 5 Gangwon-do 5
#> 6 Gwangju 5
#> 7 Jeollabuk-do 4
#> 8 Jeollanam-do 2
ggplot(cnt_province, aes(x = reorder(province, n), y = n)) +
geom_col() +
coord_flip() +
xlab("province")Dataset infomation
dataset-detailed-description.ipynb
getdata() import 3 of these datasets.
patientfrom PatientInfo.csvroutefrom PatientRoute.csvtimefrom Time.csv