Kaggle competition link.
Data, X-Ray images, are provided in *.dcm (DICOM) format. Every file contains metadata and pixel array. Since single
file load takes plenty of time (~2s) it is handy to convert them into *.jpg.
Function preprocess.convert_dcm_dataset_to_jpg could be used for dataset conversion.
It is expected such datasets exist in directories dataset_generated/train & dataset_generated/test before training.
Images in these directories should follow <SOPInstanceUID>.jpg naming convention.
- Train/Validation dataset split: 1588/150
- Model: ResnetRS50
(224, 224, 3) - No shuffle
- F1 Train score:
0.8860 - F1 Validation score:
0.8133 - Kaggle test score:
0.78058
- Epochs: 5 --> 10
- F1 Train score:
0.953 - F1 Validation score:
0.8533 - Kaggle test score:
0.79405
- Dataset (train & test) regenerated - Images with
dicom.PhotometricInterpretation == MONONCHROME2were inverted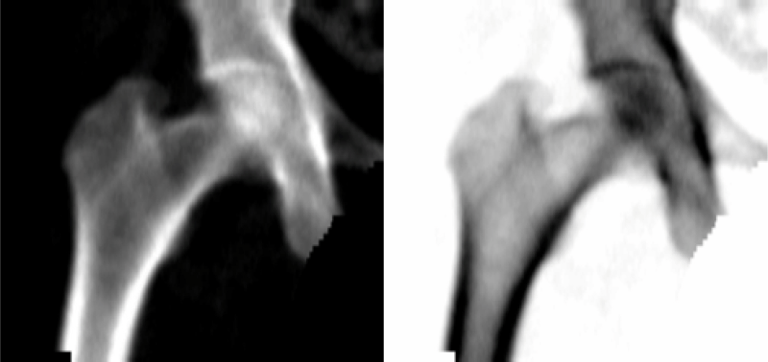
- Epochs: 8
- F1 Train score:
0.9662 - F1 Validation score:
0.8542 - Kaggle test score:
0.80246
- Keep aspect ratio of an input image and pad to model input resolution with black
- No improvement
- Simplify model architecture (just
GlobalAveragePoolingand singleDenselayer) - Increase validation dataset size to 20 % of training samples
- Shuffle training dataset after each iteration
- Batch size: 32 -> 64
- Epochs: 12
- Learning rate: 0.0001
- F1 Train score:
0.9997 - F1 Validation score:
0.8862 - Kaggle test score:
0.83277 - Reached plateau on both metrics (accuracy and F1-Score) and both datasets (train and validation)
- Use
EfficientNetV2Mwith input size(X, 320, 320, 3) - Batch size: 32
- Epochs: 12
- F1 Train score:
0.9717 - F1 Validation score:
0.9064 - Kaggle test score:
0.86083
- Multiple epochs, multiple architectures, w/o preprocessing
- No improvement, still overfitting
- Apply data augmentation to training dataset
- CLAHE, Rotate, Brightness & Contrast, (ISO/Gauss)Noise,
- Epochs: 30 (early stopped after 25)
- F1 Train score:
0.9758 - F1 Validation score:
0.9046 - Kaggle test score:
0.92087
There were 41 out of 1738 images from training dataset that were not classified correctly. Mapping between class_id
and class name is available in config.py. Incorrect labels are in format [truth]→[prediction].
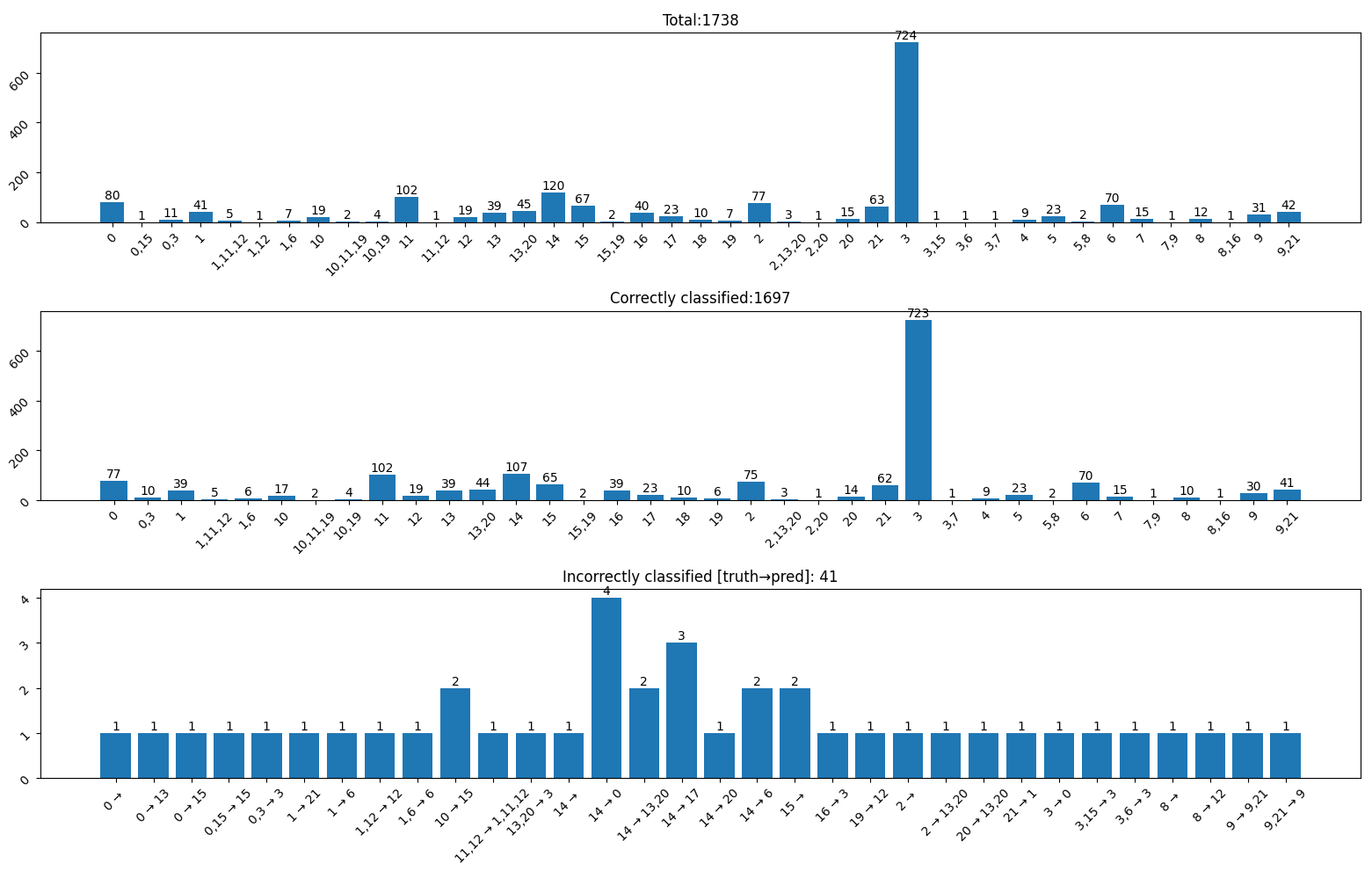
Images of 40 incorrectly classified inputs and commentary.


| Image IDs | Comment / Conclusion* |
|---|---|
0,31,32 |
Image too bright / dark / low contrast. Hard to spot features. |
1,2,7,8,13,15,17,18,20,22,23,28,34,35,38,39 |
Questionably or erroneously labeled dataset. Model's guess not bad at all. |
3,4,6,14,26,33,36 |
Truth defined as Other. Model made good guess. |
5,13,23,34 |
Highly atypical data (broken bones etc.). Model could perform better. |
10,19,35,40 |
Model found only part of the labels. |
16,21 |
Model classified most of the labels correctly, but added incorrect ones. |
11,12,24,25,27,29,30,37 |
Just wrong classification. |
13,25,36 |
Image combined from more than 1 x-ray scan. |
*Some conclusion might not be completely correct since I'm not expert in radiology.
- 25 epochs was probably too much and model was overtrained
- Epochs: 25 -> 20
- Input size:
(224, 224, 3) - F1 Train score:
0.9658 - F1 Validation score:
0.9044 - Kaggle test score:
0.93827