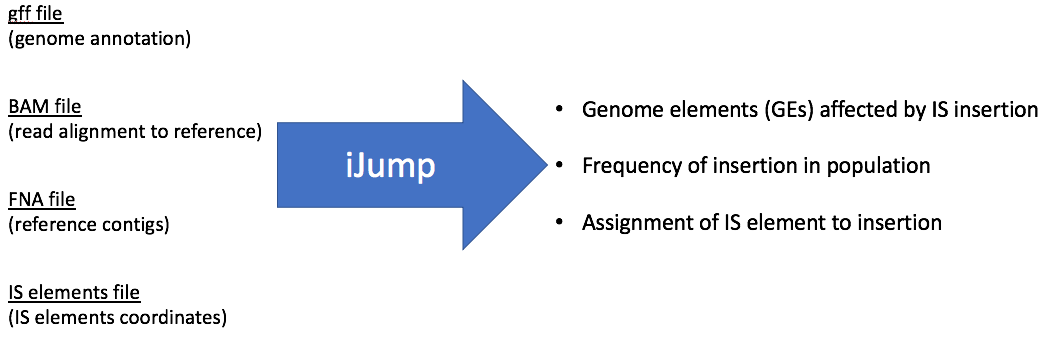
Software for search of Insertion Sequences (IS) rearrangements in evolved population sequencing data. Program is in a development stage, and I will appreciate any feedback.
iJump searches for IS elements rearrangements in evolved populations of single organism and estimates what fraction of a population is affected by the rearrangement. iJump uses soft-clipped reads to find evidense for rearrangements.
NOTE: Working with short-read-only assembled genomes is difficult with iJump. The reason is that usually IS elements are repetitive regions which are difficult to resolve for assemblers. This often result in shreading IS elements to several/many sometimes overlapped short contigs. This introduces difficulty either for boundaries determination and for mapping algorithms.
Genome rearrangements and especially IS rearrangements are powerful tools for evolution in all domains of life. Currently very few software tools exists that can work with data of mixed populations, not the clonal ones. The goal of the iJump is to estimate frequency of rearrangements in the evolved population. The scenario that was in our mind is following:
- Bacterial population is going through experimental evolution.
- During the evolution process the initial population split into several subpopulations.
- In subpopulations various IS rearrangements occur.
The only tool for bacterial population data that was found is a breseq. However although it is an excellent and comprehensive tool we've found couple limitations of its use. (1) It very slow on high coverage data and (2) it is impossible to use only selected analysis tools.
iJump do not need special installation. Just clone repository:
git clone https://github.com/sleyn/ijump.git
But it is dependent on several Python libraries:
- biopython
- pandas
- pysam
- pysamstats
- numpy
- scipy
- sklearn
The dependences could be installed in Conda environment:
conda install \
-c anaconda \
-c bioconda \
-c conda-forge \
biopython=1.79 \
pandas=1.3.5 \
numpy=1.21.0 \
scikit-learn=0.24.2 \
pysam=0.15.3 \
pysamstats=1.1.2 \
scipy=1.4.1
----IN DEVELOPMENT----
iJump requires four files for input:
- File with mobile elements coordinates
- Reference DNA contigs fasta file.
- GFF file with reference genome annotations.
- BAM file of aligned Illumina reads.
File with mobile elements coordinates shoud be tab-separated tables of the following structure:
IS_Name Contig_Name Start_position End_position
For example:
ISAcsp3 NODE_1 2980551 2981283
If you don't have file with coordinates of mobile elements you can:
- Preferred. Do manual BLAST against standalone ISFinder database. Database could be downloaded from:
- ISFinder original GitHub
- My Fork with already built BLASTn database.
Do BLASTn search:
blastn -query <Genome> -db <BLASTn database from IS.fna> -out <Output file> -outfmt 6
Parse the output table with isfinder_db_parcer.py script:
python3 isfinder_db_parcer.py -b <BLAST output in outfmt 6 format> -o <Output directory>
- Find them from ISFinder website using their BLAST against your reference contigs.
It will return you html page of hits that you can download and parse with isfinder_parser.py:
python3 isfinder_parse.py -i <ISfinder BLAST HTML page>
Both parsers will find non-overlapping hits with empirical E-value threshold 1E-30.
NOTE: It was observed that if the contig FASTA header (the line starting with ">") is long then ISFinder BLAST does not produce "Query=" string with the contig name. This line is critical for isfinder_parse.py work. If the script reports empty table please change header by using sorter contig names or removing auxiliary information.
Regular Fasta file with one Fasta-record per contig:
>Contig1
gctagctagctagctacgtagctagctagctacgtacgtacgtagcta...
>Contig2
cgtagctgctagctagctagctagcgtacgtacgtagctacgtacgta...
...
iJump is working with it's own gff module that is tuned for PATRIC/PROKKA-style GFF.
Some specifications:
-
The comment string
##sequence-region accn|[contig name] [contig start coordinate] [contig end coordinate]is required for each contig in the reference. -
Info field should have the following fields:
- ID
- product
- locus_tag
- (optional, if gene has trivial name) gene
Example:
##gff-version 3
##sequence-region accn|NODE_1_length_3909467_cov_533.478_ID_22129 1 3909467
NODE_1_length_3909467_cov_533.478_ID_22129 FIG CDS 34 336 . - 1 ID=fig|400667.82.peg.1;product=hypothetical protein;locus_tag=AUO97b_00141
NODE_1_length_3909467_cov_533.478_ID_22129 FIG CDS 352 1578 . - 1 ID=fig|400667.82.peg.2;product=phage replication protein Cri;locus_tag=AUO97b_00142;gene=cri
NODE_1_length_3909467_cov_533.478_ID_22129 FIG CDS 1724 2098 . + 2 ID=fig|400667.82.peg.3;product=helix-turn-helix family protein;locus_tag=AUO97b_00143
If you have another style of GFF unfortunately you have to reformat it at the current stage of development.
BAM file with aligned short reads. BAM file should contain soft-slipped reads. On the current stage iJump don't use hard-clipped reads.
NOTE: If you have aligner (like BWA-mem) that produses both soft- and hard-clipped reads you should use option to use only soft-clipped reads or multiply frequency assessments by 2.
iJump has two workflows:
- "Average". iJump estimates frequency of IS element insertions in genes or intergenic region based on average coverage of the gene. This workflow is original and made to estimate the case of very heterogeneous populations. This workflow has low accuracy. However it has a good sensistivity.
- "Precise". iJump will try to localize insertion coordinates first and then estimate frequency of insertion in population. Less tested but logically more correct.
Example of "Average" workflow:
python3 ijump.py \
-a Sample.bam \
-r Escherichia_coli_BW25113.fna \
-g Escherichia_coli_BW25113.gff \
-i ISTable_processing.txt \
-o average_out
Example of "Precise" workflow:
python3 ijump.py \
-a Sample.bam \
-r Escherichia_coli_BW25113.fna \
-g Escherichia_coli_BW25113.gff \
-i ISTable_processing.txt \
--estimation_mode precise \
-o precise_out
Available parameters:
usage: ijump.py [-h] [-a ALN] [-r REF] [-g GFF] [-i ISEL] [-c] [-o OUTDIR]
[-w WD] [--radius RADIUS] [--estimation_mode ESTIMATION_MODE]
[--version]
iJump searches for small frequency IS elements rearrangements in evolved
populations
optional arguments:
-h, --help show this help message and exit
-a ALN, --aln ALN BAM or SAM alignment file
-r REF, --ref REF Reference genome in FASTA format
-g GFF, --gff GFF Annotations in GFF format for reference genome.
Required for average mode.
-i ISEL, --isel ISEL File with IS elements coordinates
-c, --circos Set flag to build input files for CIRCOS
-o OUTDIR, --outdir OUTDIR
Output directory. Default: . (current)
-w WD, --wd WD Work directory. Default: ijump_wd (current)
--radius RADIUS Radius around IS elements boundaries to search soft
clipped reads.
--estimation_mode ESTIMATION_MODE
Specifies how the IS frequency will be esimated.
'average' - by averaging the region coverage and
number of clipped reads. Or 'precise' - iJump will try
to separate each insertion event.
--version Print iJump version and exit.
If you have several related samples and want to compare them side by side you can copy all ijump_report_by_is_reg.txt files in one folder, rename them as ijump_<Sample name>.txt* and run:
python3 combibe_results.py -d [Folder with ijump report files] -o [Output file with the combined table] -g [GFF file. If provided will add functional annotation of the region]
This will merge all results in one table.
Available parameters:
python3 combine_results.py -h
usage: combine_results.py [-h] [-d DIR_REPORT] [-o OUTPUT] [-g [GFF]]
[-p PREFIX] [-m IJUMP_MODE] [--lab_format]
[--clonal] [-a [A_SAMPLES]]
[--precise_mode PRECISE_MODE]
Tool that combines ijump reports from several files into one summary table
optional arguments:
-h, --help show this help message and exit
-d DIR_REPORT, --dir_report DIR_REPORT
Directory with report files
-o OUTPUT, --output OUTPUT
Output table file
-g [GFF], --gff [GFF]
Path to gff file
-p PREFIX, --prefix PREFIX
If set would be used as prefix
-m IJUMP_MODE, --ijump_mode IJUMP_MODE
Variant of iiJump pipeline: "average" or "precise".
Default: "average"
--lab_format If set, output internal laboratory
--clonal If set, runs clonal merging
-a [A_SAMPLES], --a_samples [A_SAMPLES]
Path to folder with unevolved population samples. Used
for clonal analysis.
--precise_mode PRECISE_MODE
If "dense" mode selected iJump will try to combine and
sum observations based on one edge. For example: If
observation 1 has coordinates 100-105 and other 100-0
(right junction is undefined) the results will be
combined. The options are "dense" and "sparse".
Default: dense