Source code for our paper "Shape-Aware Complementary-Task Learning for Multi-Organ Segmentation" as described in Early access on 10th International Workshop on Machine Learning in Medical Imaging (MLMI 2019), MICCAI 2019).
Authors: Fernando Navarro, Suprosanna Shit, Ivan Ezhov, Johannes C. Paetzold, Andrei Gafita, Jan Peeken , Stephanie E. Combs, and Bjoern H. Menze.
You need to have following in order for this library to work as expected
- python >= 3.6.5
- pip >= 18.1
- tensorflow-gpu = 1.9.0
- tensofboard = 1.9.0
- numpy >= 1.15.0
- dipy >= 0.14.0
- matplotlib>= 2.2.2
- nibabel >= 1.15.0
- pandas >= 0.23.4
- scikit-image >= 0.14.0
- scikit-learn >= 0.20.0
- scipy >= 1.1.0
- seaborn >= 0.9.0
- SimpleITK >= 1.1.0
- tabulate >= 0.8.2
- xlrd >= 1.1.0
Run pip install -r requirements.txt
Request access to Visceral Anatomy 3 data Anatomy3.
Run the python script data2tfrecords.py, make sure you change the paths to data_folder and tf_file. The data should be in the following format:
└── data folder
├── 10000004_1
├── 10000004_1_CT_wb.nii.gz
├── 10000004_1_CT_wb_seg.nii.gz
├── 10000007_1
├── 10000004_1_CT_wb.nii.gz
├── 10000004_1_CT_wb_seg.nii.gz
| .......................
You need to generate two files; a training and a validation tfrecord file. Change excel_file and other variables accordingly.
Run the python script train_val.py. Make sure to change file paths for tfrecord files according to your configuration.
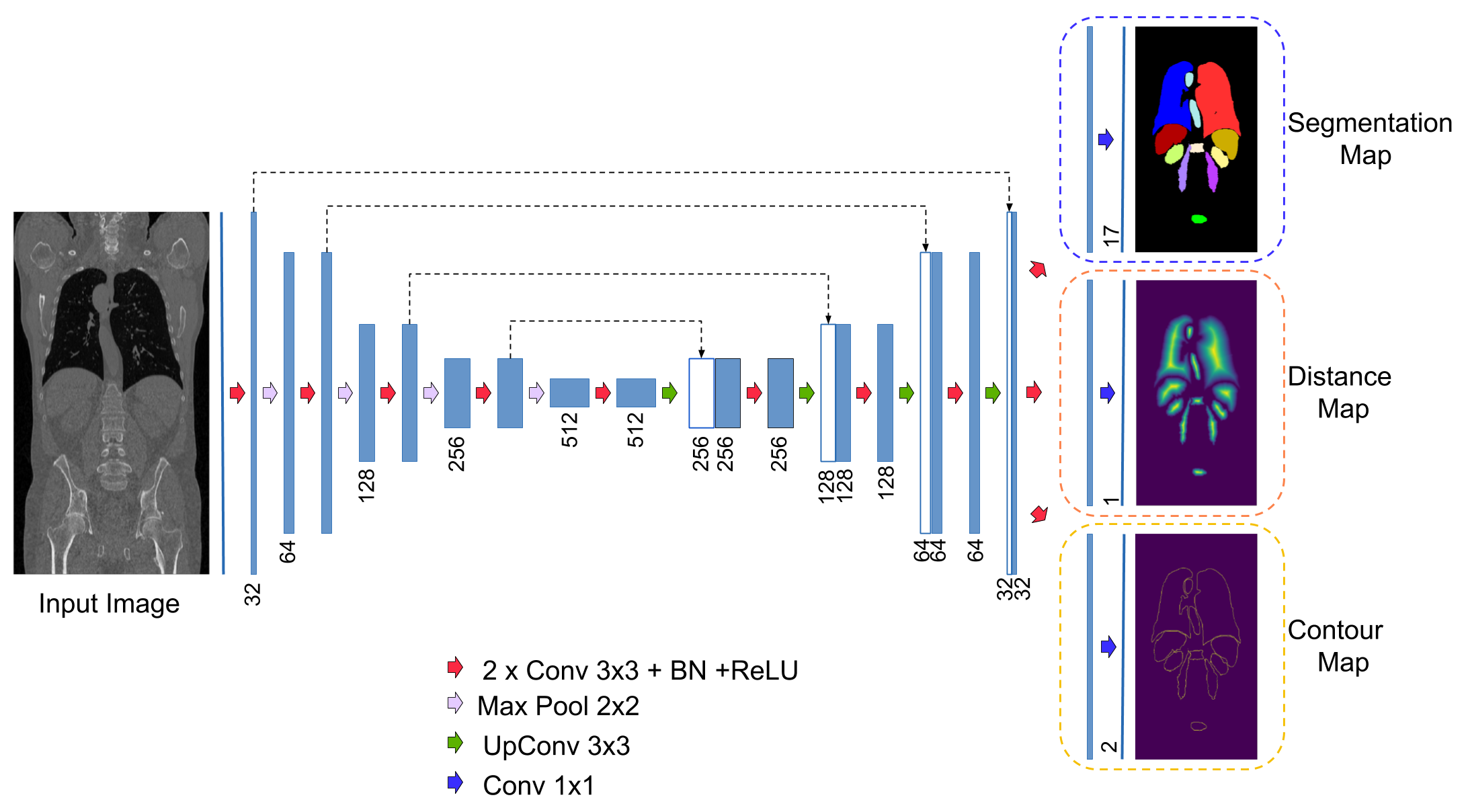
Enable segmentation branch, countour branch and distance branch by changing the variable branches=['DistanceTransformBranch','EdgesBranch', 'SegBranch']. branches=['SegBranch'] means only the segmenation branch is activated.
Run the python script inference.py. Follow the commends in the script to change variables according to your training model and file paths.
Please cite our paper if it is useful for your research:
@article{navarro2019shape,
title={Shape-Aware Complementary-Task Learning for Multi-Organ Segmentation},
author={Navarro, Fernando and Shit, Suprosanna and Ezhov, Ivan and Paetzold, Johannes and Gafita, Andrei and Peeken, Jan and Combs, Stephanie and Menze, Bjoern},
journal={arXiv preprint arXiv:1908.05099},
year={2019}
}
- Fernando Navarro - ferchonavarro
- Suprosanna Shit - suprosanna
Let us know if you face any issues. You are always welcome to report new issues and bugs and also suggest further improvements. And if you like our work hit that start button on top. Enjoy :)