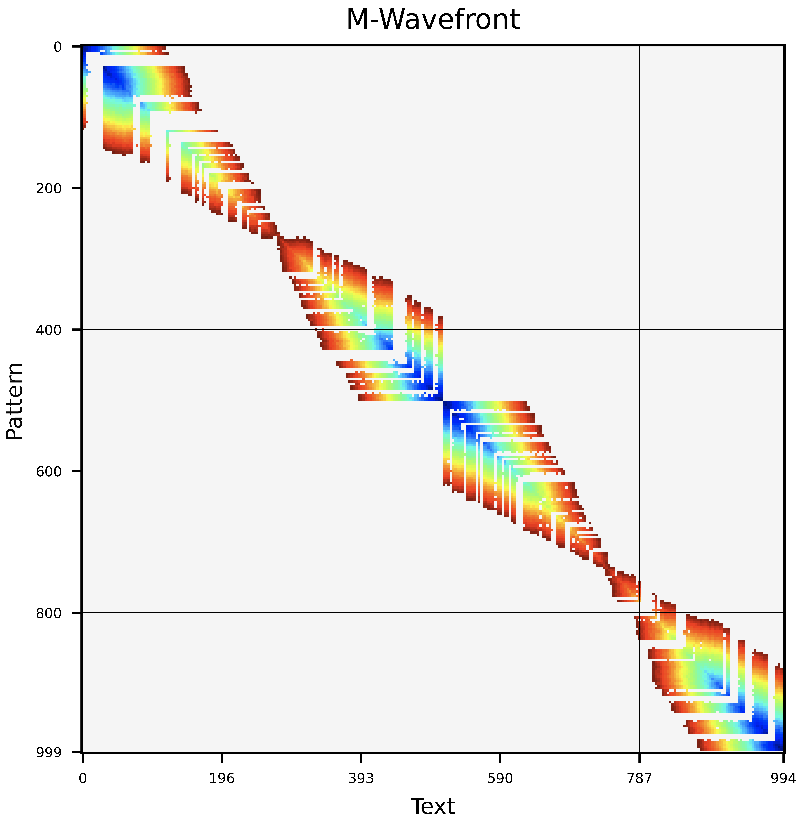
This repository is a WFA2-lib development branch implementing the bidirectional WFA (BiWFA) algorithm capable of O(ns) alignment using O(s) memory.
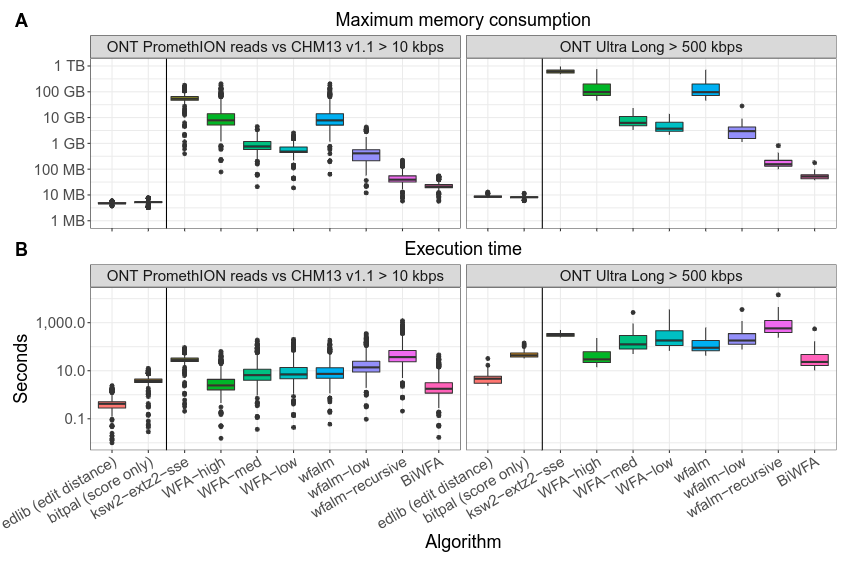
BiWFA is the first gap-affine algorithm capable of computing optimal alignments in O(s) memory while retaining the WFA's time complexity of O(ns). The core idea of the BiWFA algorithm is to perform the WFA algorithm simultaneously in both directions on the strings: from start to end, and from end to start. Each direction will only retain max{x,o+e} wavefronts in memory. This is insufficient to perform a full traceback. However, when they "meet" in the middle, we can infer a breakpoint in the alignment that divides the optimal score roughly in half. Then, we can then apply the same procedure on the two sides of the breakpoint recursively. In practice, our implementation never requires more than a few hundred MBs aligning noisy Oxford Nanopore Technologies reads up to 1 Mbp long while maintaining competitive execution times.
Usage: Same as WFA2-lib
./bin/align_benchmark -i sequences.seq -o out.alg --affine-penalties 0,3,6,1
For more documentation on the library and other features, go to WFA2-lib.
-
A note for the fierce competitors. I can understand that science and publishing have become a fierce competition these days. Many researchers want their methods to be successful and popular, seeking funding, tenure, or even fame. If you are going to benchmark the WFA using the least favourable configuration, careless programming, and a disadvantageous setup, please, go ahead. But remember, researchers like you have put a lot of effort into developing the WFA. We all joined this "competition" because we sought to find better methods that could truly help other researchers. So, try to be nice, tone down the marketing, and produce fair evaluations and honest publications.
-
We develop methods to help the community and push science forward. If you like the BiWFA ideas and would like to use it, feel free to copy, improve, and even reach out and request more features. In return, we just ask for proper acknowledgement, citation, or comment on where the methods and ideas came from.
Git clone and compile the library, tools, and examples.
git clone https://github.com/smarco/BiWFA-paper
cd BiWFA-paper
make clean all
The BiWFA-paper includes the benchmarking tool align-benchmark to test the BiWFA. This tool takes as input a dataset containing pairs of sequences (i.e., pattern and text) to align. Patterns are preceded by the '>' symbol and texts by the '<' symbol. Example:
>ATTGGAAAATAGGATTGGGGTTTGTTTATATTTGGGTTGAGGGATGTCCCACCTTCGTCGTCCTTACGTTTCCGGAAGGGAGTGGTTAGCTCGAAGCCCA
<GATTGGAAAATAGGATGGGGTTTGTTTATATTTGGGTTGAGGGATGTCCCACCTTGTCGTCCTTACGTTTCCGGAAGGGAGTGGTTGCTCGAAGCCCA
>CCGTAGAGTTAGACACTCGACCGTGGTGAATCCGCGACCACCGCTTTGACGGGCGCTCTACGGTATCCCGCGATTTGTGTACGTGAAGCAGTGATTAAAC
<CCTAGAGTTAGACACTCGACCGTGGTGAATCCGCGATCTACCGCTTTGACGGGCGCTCTACGGTATCCCGCGATTTGTGTACGTGAAGCGAGTGATTAAAC
[...]
Once you have the dataset ready, you can run the align-benchmark tool to benchmark the performance:
$> ./bin/align_benchmark -i sample.dataset.seq
...processed 10000 reads (benchmark=125804.398 reads/s;alignment=188049.469 reads/s)
...processed 20000 reads (benchmark=117722.406 reads/s;alignment=180925.031 reads/s)
[...]
...processed 5000000 reads (benchmark=113844.039 reads/s;alignment=177325.281 reads/s)
[Benchmark]
=> Total.reads 5000000
=> Time.Benchmark 43.92 s ( 1 call, 43.92 s/call {min43.92s,Max43.92s})
=> Time.Alignment 28.20 s ( 64.20 %) ( 5 Mcalls, 5.64 us/call {min438ns,Max47.05ms})
The align-benchmark tool will finish and report overall benchmark time (including reading the input, setup, checking, etc.) and the time taken by the algorithm (i.e., Time.Alignment). If you want to measure the accuracy of the alignment method, you can add the option --check and all the alignments will be verified.
$> ./bin/align_benchmark -i sample.dataset.seq --check
...processed 10000 reads (benchmark=14596.232 reads/s;alignment=201373.984 reads/s)
...processed 20000 reads (benchmark=13807.268 reads/s;alignment=194224.922 reads/s)
[...]
...processed 5000000 reads (benchmark=10625.568 reads/s;alignment=131371.703 reads/s)
[Benchmark]
=> Total.reads 5000000
=> Time.Benchmark 7.84 m ( 1 call, 470.56 s/call {min470.56s,Max470.56s})
=> Time.Alignment 28.06 s ( 5.9 %) ( 5 Mcalls, 5.61 us/call {min424ns,Max73.61ms})
[Accuracy]
=> Alignments.Correct 5.00 Malg (100.00 %) (samples=5M{mean1.00,min1.00,Max1.00,Var0.00,StdDev0.00)}
=> Score.Correct 5.00 Malg (100.00 %) (samples=5M{mean1.00,min1.00,Max1.00,Var0.00,StdDev0.00)}
=> Score.Total 147.01 Mscore uds. (samples=5M{mean29.40,min0.00,Max40.00,Var37.00,StdDev6.00)}
=> Score.Diff 0.00 score uds. ( 0.00 %) (samples=0,--n/a--)}
=> CIGAR.Correct 0.00 alg ( 0.00 %) (samples=0,--n/a--)}
=> CIGAR.Matches 484.76 Mbases ( 96.95 %) (samples=484M{mean1.00,min1.00,Max1.00,Var0.00,StdDev0.00)}
=> CIGAR.Mismatches 7.77 Mbases ( 1.55 %) (samples=7M{mean1.00,min1.00,Max1.00,Var0.00,StdDev0.00)}
=> CIGAR.Insertions 7.47 Mbases ( 1.49 %) (samples=7M{mean1.00,min1.00,Max1.00,Var0.00,StdDev0.00)}
=> CIGAR.Deletions 7.47 Mbases ( 1.49 %) (samples=7M{mean1.00,min1.00,Max1.00,Var0.00,StdDev0.00)}
Using the --check option, the tool will report Alignments.Correct (i.e., total alignments that are correct, not necessarily optimal), and Score.Correct (i.e., total alignments that have the optimal score).
Note that the overall benchmark time will increase due to the overhead introduced by the checking routine, however the Time.Alignment should remain the same.
Feedback and bug reporting is highly appreciated. Please report any issue or suggestion on github or email to the main developer (santiagomsola@gmail.com).
BiWFA is distributed under MIT licence.
Santiago Marco-Sola, Jordan M Eizenga, Andrea Guarracino, Benedict Paten, Erik Garrison, Miquel Moreto. "Optimal gap-affine alignment in O(s) space". Bioinformatics, 2023.