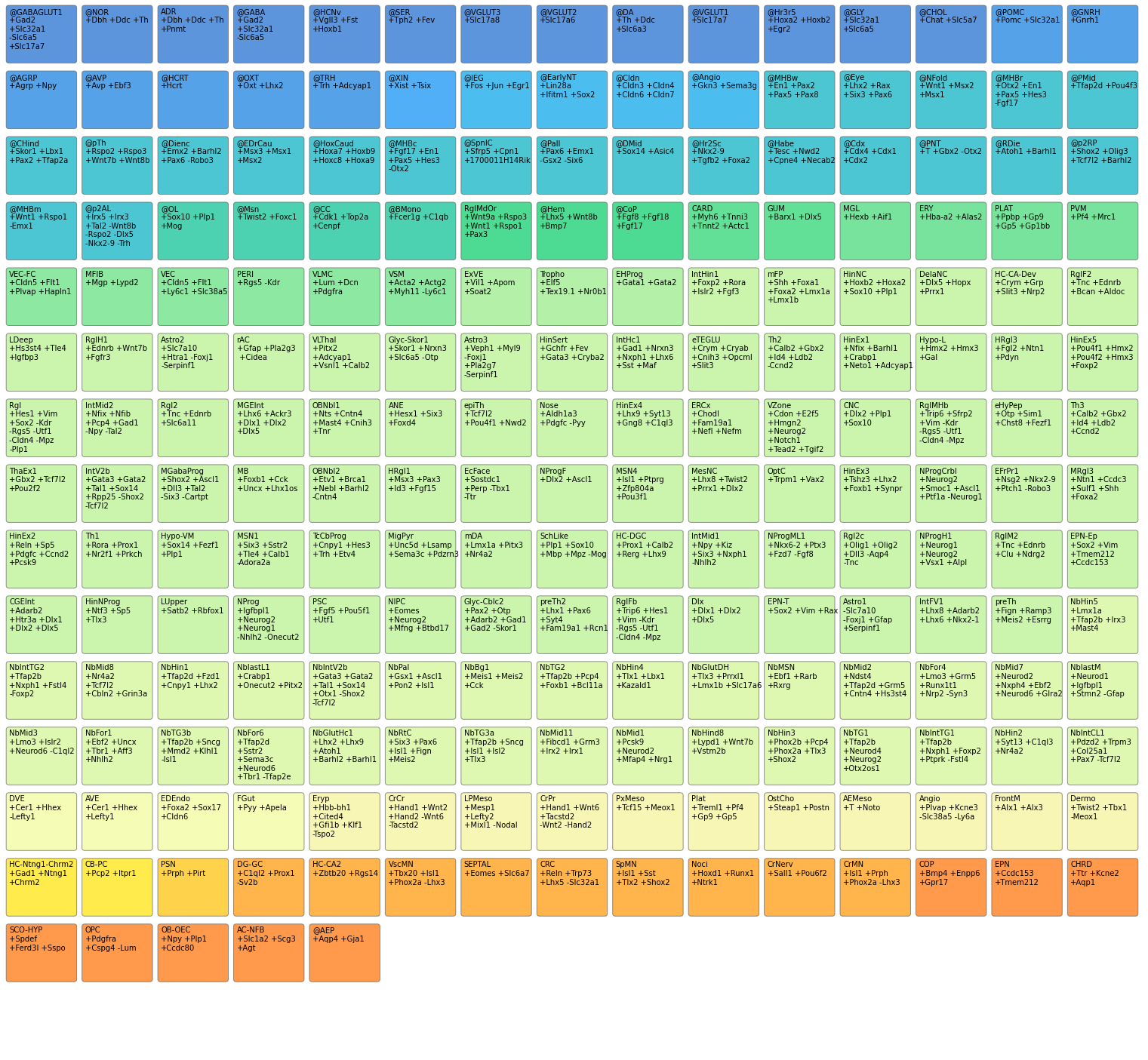
Computer-readable annotation for developmental cell types and states in the developing mouse brain.
This resource has been produced as a part of the research related to the paper:
Molecular Architecture of the Developing Mouse Brain
Gioele La Manno*+, Kimberly Siletti*, Alessandro Furlan, Daniel Gyllborg, Elin Vinsland, Christoffer Mattsson Langseth, Irina Khven, Anna Johnsson, Mats Nilsson, Peter Lönnerberg, Sten Linnarsson+
This repository contains a database of auto-annotation labels that were constructed cross-referencing literature and observation from the data. The auto-annotation tags can be used for data exploration and to manipulate a big dataset in a semi-automatic way.
Cell type/state labels are defined as text documents, one per label, with a short computer-readable section as well as a free-form section. The former, exemplified below, consists of four mandatory fields: a name, an abbreviation, a definition and a category field; the latter is used to provide context, justification and references, increasing the usefulness of annotation labels.
name: Cranial Neural Crest Cell
abbreviation: CNC
definition: +Dlx2 +Plp1 +Sox10
categories: Ectodermal NeuralCrest FaceThe abbreviation and name are unique identifiers and they are simply for human consumption. The categories field contains attributes that can be used to group the labels in meta-groups. The definition is the most important field and effectively represents the label assignment rule. It consists of a list of gene names, with the name prefixed with “+” if the gene is required to be present or “-” if required to be absent. To determine if a label can be attributed to a particular cluster we use a statistical binarization. The condition for labeling the cluster is met when all the “+” genes are binarized as “on” and all the “-” genes are binarized as “off”.
We constructed the definitions so that they apply globally. That is, each label was intended to mark any cell in any part of the body that conforms to the definition. Considering the vast number of genes available and the different combinations of those, different definitions for the same label could be possible. Here we favored definitions that consisted of a short list of genes, of which as many as possible are well known genes whose function is described in literature. The reasoning behind that being that those are most robust as they tend to remain valid when applied to different datasets.
The auto-annotation labels can be applied downstream to any statistical text that determines whether a gene is robustly expressed in a pool of cells or not in a dihcotomic way. In La Manno et al. 2020 for example we used the Bayesian Trinarization procedure implemented in cytograph.