Single cell RNA sequencing allows us to interrogate the expression of thousands of genes at single cell resolution. The rapid growth of scRNA-seq data has also created an unique set of challenges: how do we visualize and extract useful biological information from the massive and high-dimensional scRNA-seq data?
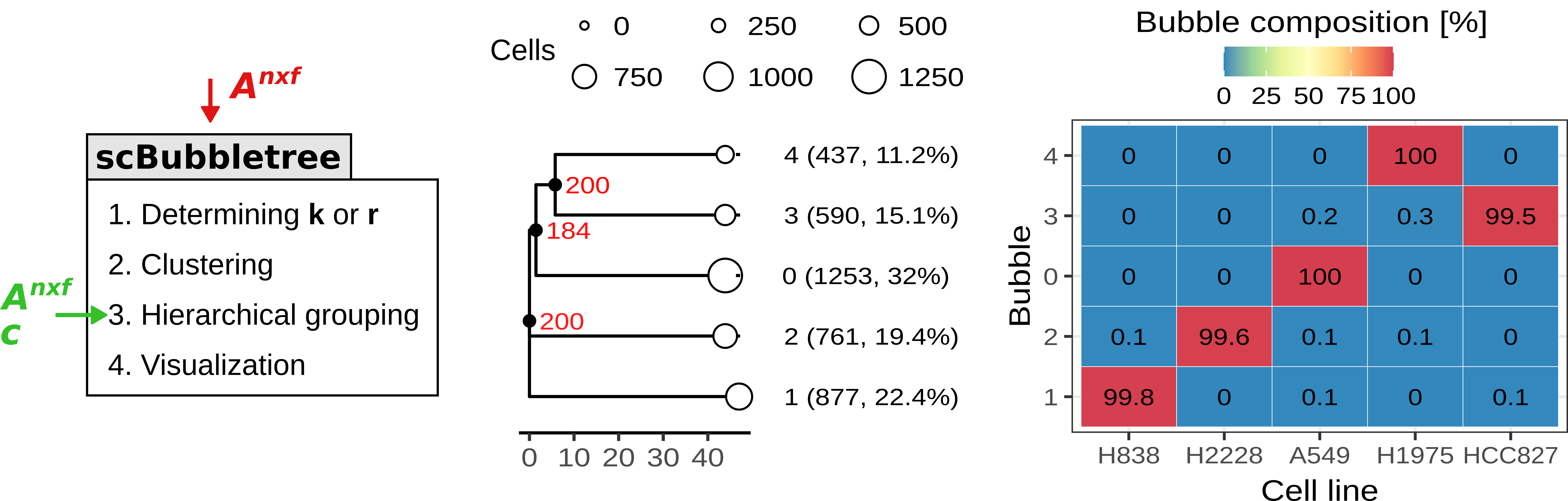
scBubbletree is a scalable method for visualization of scRNA-seq data. The method identifies clusters of cells of similar transcriptomes and visualizes such clusters as “bubbles” at the tips of dendrograms (bubble trees), corresponding to quantitative summaries of cluster properties and relationships. scBubbletree stacks bubble trees with further cluster-associated information in a visually easily accessible way, thus facilitating quantitative assessment and biological interpretation of scRNA-seq data.
scBubbletree is an R-package available from Bioconductor:
https://bioconductor.org/packages/scBubbletree/
To install this package, start R and enter:
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("scBubbletree")Case studies are provided in the directory /vignettes