Samples
Diabetes : 3 samples
Control : 3 Samples
Platform
Nanostring Ncounter Platform
Probes
Endogeneous : 798
HouseKeeping : 6
Ligation : 6
Expression data was normalized using Nsolver Software
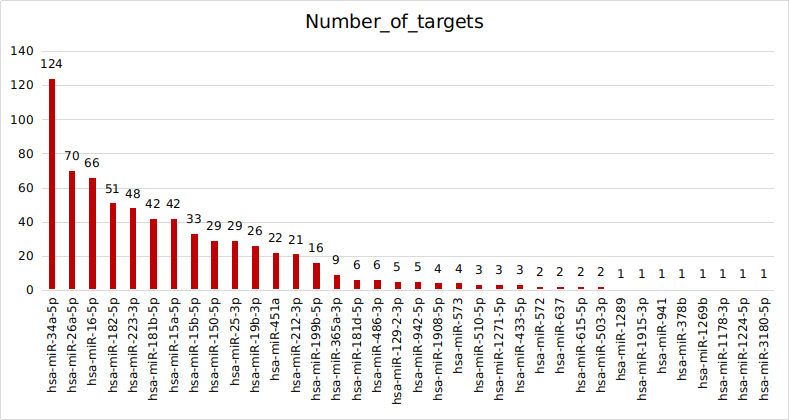
DEmiRNA : 51
Fold Change : 1.5
P-value : <= 0.05
DEmiRNA : 15
Fold Change : 2
P-value : <= 0.05
Experimentally Validated MTI was retrived from MiRTarBase
Experiments
Luciferase reporter assay
ELISA
Reporter assay
Flow
Western blot
qRT-PCR
Luciferase assay
Immunofluorescence
GFP reporter assay
Immunohistochemistry
Immunocytochemistry
In situ hybridization
Northern blot
Immunoprecipitaion
Immunoblot
Microarray
FACS
Chromatin immunoprecipitation
ChIP-seq
Proteomics
Flow cytometry
ChIP
EMSA
Support Type
Functional MTI
Functional MTI (Weak)
Non-Functional MTI
Non-Functional MTI (Weak)
Considered only Functional MTI from the Support Type
The targets of DEmiRNAs for given for functional enrichment.
The ontologies were further filtered with keywords:
insulin
glucagon
glucose
lipid