Note: This is all pre-alpha stuff (i.e. being worked on extensively, there will be breaking changes, the repo may be burnt down and rebuilt at any time). Extensive documentation will be made available at a later date when this is ready for general use.
Documentation can be found here: https://socialgene.github.io
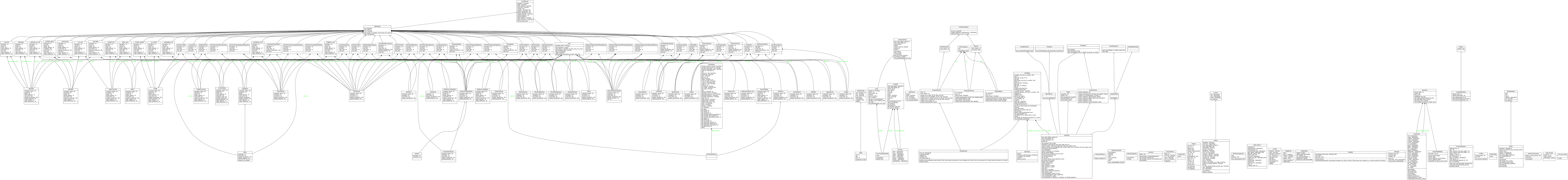
Design
The code is organized under a number of submodules/directories:
- base: core functions of the library
- cli: all command line interface code
- clustermap: used to convert a socialgene object to clustermap json
- findmybgc
- hashing
- hmm: code for working with HMMER
- neo4j: code for working with SocialGene Neo4j databases
- parsers: external file parsers (e.g. genbank, fasta, HMMER results, etc)
- scoring: functions for measuring protein similarity
- taxonomy
- utils
Installation with pip
https://pypi.org/project/socialgene
pip install socialgeneCreate conda environment and install python package inside
git clone https://github.com/socialgene/sgpy.git
cd sgpy
make create_condaBuild Python package from source
git clone https://github.com/socialgene/sgpy.git
cd sgpy
make install_pythonBuild local Docker image
git clone https://github.com/socialgene/sgpy.git
cd sgpy
make build_docker_imageRun pytest tests
git clone https://github.com/socialgene/sgpy.git
cd sgpy
make create_conda
make pytestRun all tests
git clone https://github.com/socialgene/sgpy.git
cd sgpy
make create_conda
make run_ciUser-facing classes
SocialGene()
This is the main class that most other user-facing classes should/do inherit from
FindMyBGC()
SingleProteinSearch()
Common example use cases
Starting with a single input protein and
Starting with a set of proteins (BGC) and
Other
Most of the the classes that describe the structure of SocialGene() (e.g. proteins, domains, loci) live in socialgene/src/socialgene/classes/molbio.py