cluster graph(s) with the leiden algorithm
This is a wrapper script around vtraag's Leiden
Algorithm[1]. Put this script in your path,
and then you can call it from anywhere!
# get a graph
wget https://github.com/twesleyb/leiden/raw/master/data/adjm.zip
unzip adjm.zip && cd adjm/
# cluster the graph with the leiden algorithm and surprise
leiden adjm.csv -q Surprise
# or try the CPM algorithm
leiden adjm.csv -q CPM -g 1
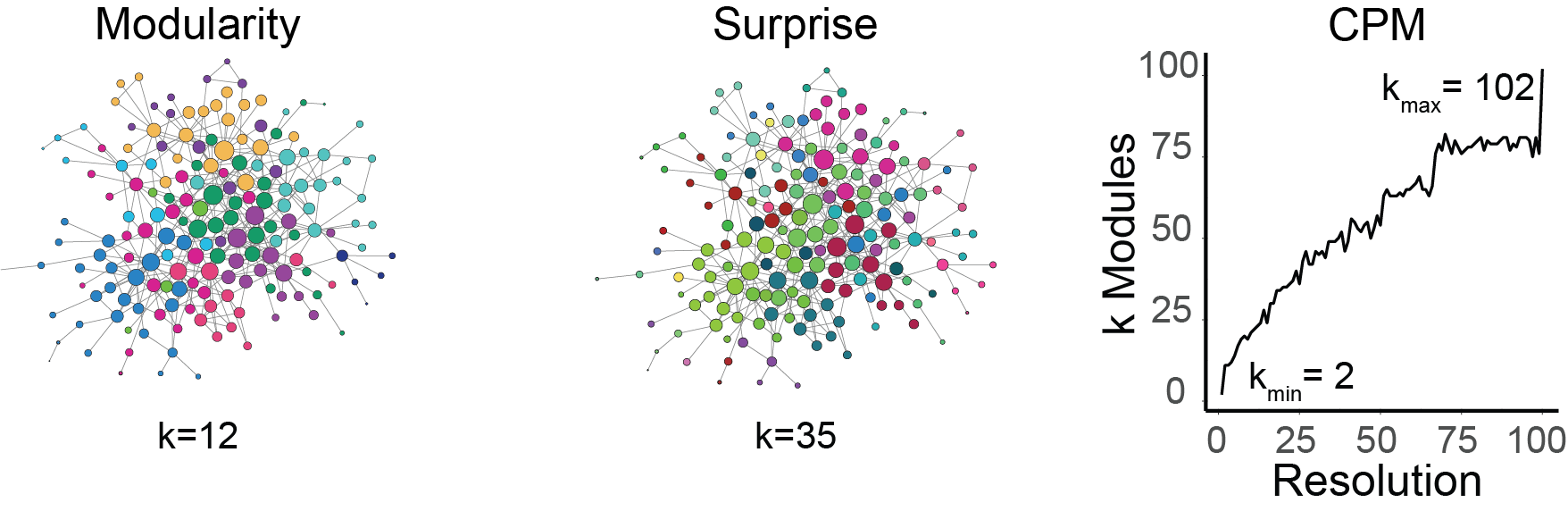
# NOTE: currently only supports a single resolution!Clustering of the iPSD protein-protein interaction graph from Uezu2016[2] using three different leidenalg methods, Modularity, Surprise, and CPM. The networks are visualized using Cytoscape.
I recommend using a virtual environment such as conda. Then insure you have
installed the following python dependencies:
- leidenalg
- igraph
conda install -c conda-forge igraph leidenalg
[1] From Louvain to Leiden: guaranteeing well-connected communities.
Traag, V.A., Waltman. L., Van Eck, N.-J. (2018). Scientific reports, 9(1), 5233.
DOI: 10.1038/s41598-019-41695-z
[2] Identification of an Elaborate Complex Mediating Postsynaptic Inhibition.
Akiyoshi Uezu, Daniel J. Kanak, Tyler W.A. Bradshaw, Erik J. Soderblom,
Christina M. Catavero, Alain C. Burette, Richard J. Weinberg, and Scott H. Soderling.
Science, 2016 Sep 9; 353(6304): 1123–1129.
DOI: 10.1126/science.aag0821