Type Inference Evaluation Scripts & Accessory Apps (used for the StaTIX benchmarking)
\authors: (c) Soheil Roshankish, Artem Lutov artem@exascale.info
\license: Apache License, Version 2.0
\organization: eXascale Infolab
\date: 2017-08
Scripts and accessory Java applications used for the type inference benchmarking of StaTIX.
evalgt.sh- executes the evaluation app (xmeasures, gecmi, etc) specified number of times with the specified options on each*.cnlfile in each specified input directory, evaluating against the _gt.cnl ground-truth.shufrdfs.sh- shuffles and reduces input RDF dataset in N3 format to the specified ratio.execfile.sh- executes commands from the specified file tracing the resource consumption.mkevaldirs.sh- creates directories corresponding to the ground-truth files to put evaluating type inference results there.linkfiles.sh- links type inference (clustering) results of the algorithm(s) to the corresponding evaluating directories made bymkevaldirs.sh.
The scripts require any POSIX compatible execution environment (Linux or Unix). Java apps were tested on Java 1.8+.
To perform batch execution of the clustering algorithms copy / unpack the required algorithm to the dedicated directory <algdir> and link / copy there scripts/execfile.sh, scripts/<algname>.exs, exectime. Update paths in the <algname>.exs if required.
Run the batch execution: ./execfile.sh <algname>.exs. It produces evals_<algname>.rcp containing execution time and memory consumption measurements in the current directory and resulting clusterings (type inference) according to the specified parameters in the <algname>.exs.
To perform batch evaluation of the type inference accuracy, create a dedicated directory <evals>/ and link / copy there scripts/evalgt.sh, scripts/linkfiles.sh, scripts/mkevaldirs.sh, xmeasures, gecmi and ground-truth files from the data/<datasets>_gt (or produce them). The ground-truth files contain for each #type property space separated subject ids, sequentially enumerated from 0. Run ./mkevaldirs.sh to create directories that will hold type inference results to be evaluated against the respective ground-truth files. Then run linkfiles.sh <results_dir> to link algorithm(s) results to the corresponding directories to be evaluated. And finally run the batch evaluations using evalgt.sh script. See evalgt.sh -h for details. It produces the required evaluations (eval_<evalapp-params>.txt files) in the current directory, which are the accuracy results of the type inference.
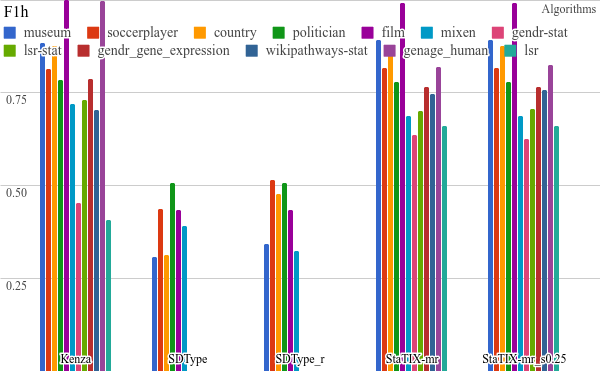
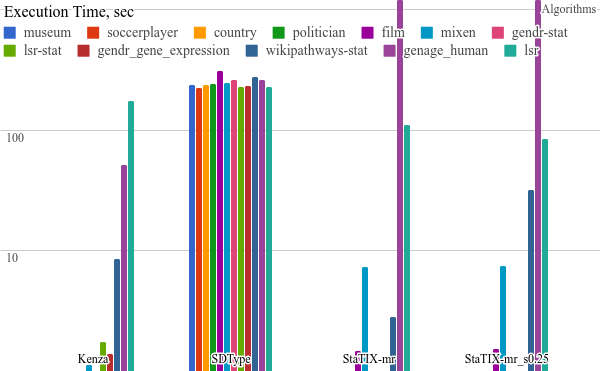
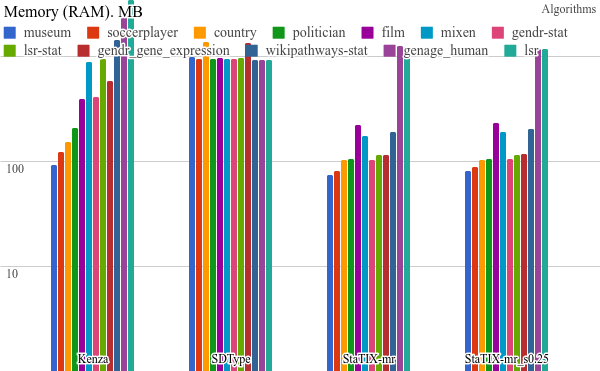
All the benchmarking results) are available online and some plots are listed below. StaTIX executed with -m, -f -r m parameters in non-supervised (StaTIX-mr) and semi-supervised (StaTIX-mr_s0.25) modes.
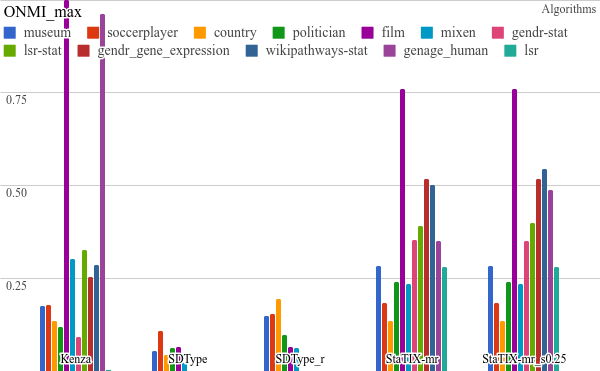 Higher is better.
Higher is better.
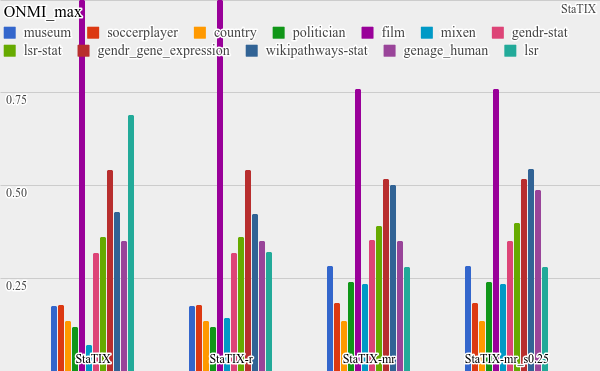 Higher is better.
Higher is better.
Note: Please, star this project if you use it.
- StaTIX - Statistical Type Inference (both fully automatic and semi supervised) for RDF datasets in N3 format.
- xmeasures - Extrinsic clustering measures evaluation for the multi-resolution clustering with overlaps (covers): F1_gm for overlapping multi-resolution clusterings with possible unequal node base and standard NMI for non-overlapping clustering on a single resolution.
- OvpNMI - NMI evaluation method for the overlapping clusters (communities) that is not compatible with the standard NMI value unlike GenConvNMI, but it is much faster than GenConvNMI.
- GenConvNMI - Overlapping NMI evaluation that is (unlike
onmi) compatible with the original NMI and suitable for both overlapping and multi resolution (hierarchical) clusterings. - ExecTime - A lightweight resource consumption profiler.
- PyCABeM - Python Benchmarking Framework for the Clustering Algorithms Evaluation. Uses extrinsic (NMIs) and intrinsic (Q) measures for the clusters quality evaluation considering overlaps (nodes membership by multiple clusters).