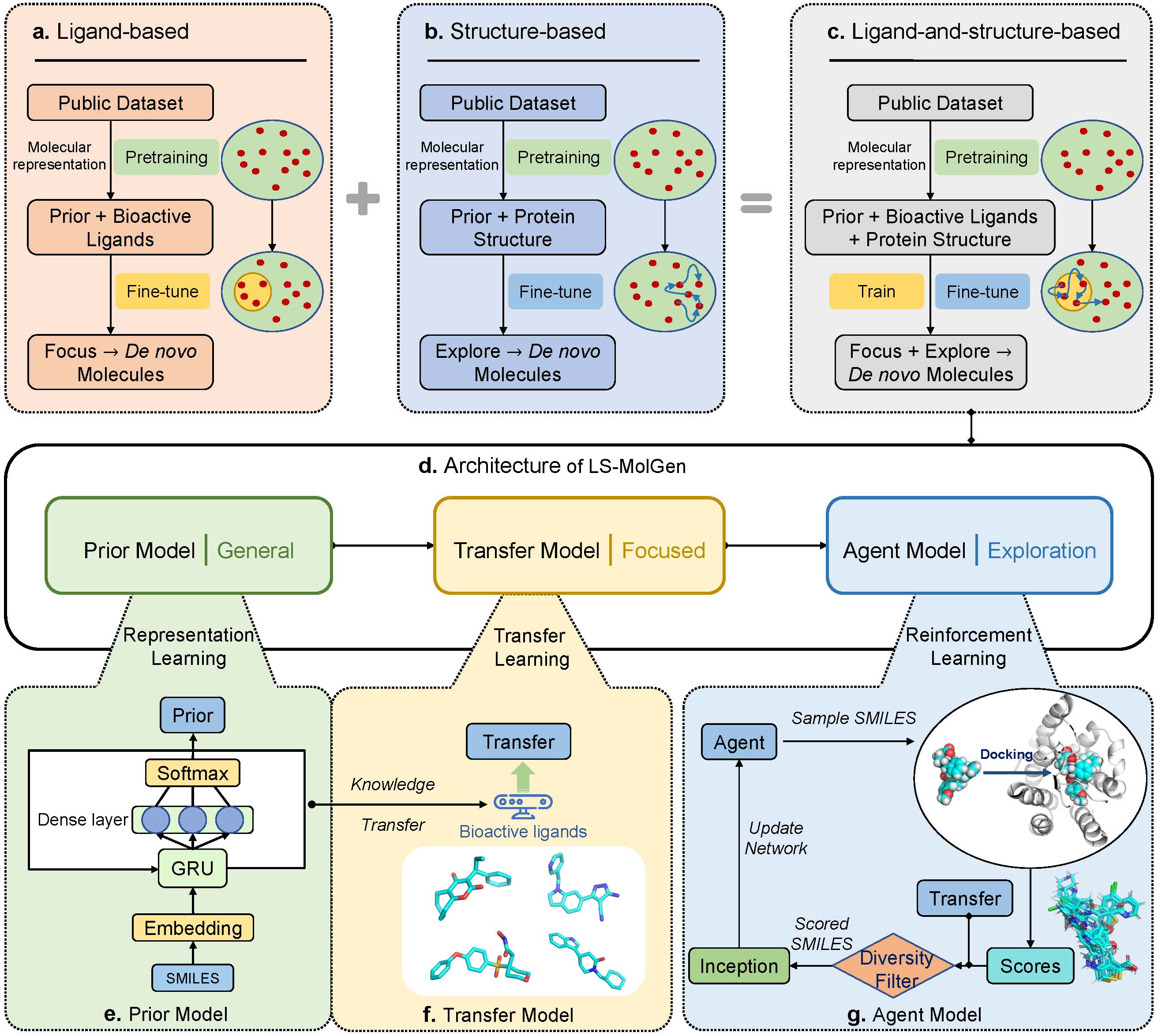
A ligand-and-structure dual-driven deep reinforcement learning method for target-specific molecular generation.
Paper: https://pubs.acs.org/doi/10.1021/acs.jcim.3c00587
You can use the environment.yml file to create a new conda environment with all the necessary dependencies for LS-MolGen:
git clone https://github.com/songleee/LS-MolGen.git
cd LS-MolGen
conda env create -f environment.yml
conda activate LS-MolGen
To run this code, especially the reinforcement learning part, the following softwares are required:
-
LeDock: the program used to get the docking score. Download from here. -
Open Babel: the software used to prepare the ligand. Download from here.
After download the release packages of them, remember to add the PATH of them to the environment variables.
LS-MolGen includes three sub-modules:
-
pre_train.py: Used for pre-training on big dataset. -
transfer_learning.py: Used for fine-tuning the pre-trained neural network on a small dataset of molecules with known bioactivity. -
reinforcement_learning.py: Used for generating new molecules with high affinity and novelty using the reinforcement learning algorithm.
Example of running the command:
python pre_train.py
python transfer_learning.py
python reinforcement_learning.py
LS-MolGen was developed by Song Li as part of a research project at Shanghai Jiao Tong University.
LS-MolGen is released under the Apache License. See the LICENSE file for details.