/ˈviːteɪ/
The vitae package makes creating and maintaining a Résumé or CV with R Markdown simple. It provides a collection of LaTeX and HTML templates, with helpful functions to add content to the documents. There are currently 6 templates available in this package:
| vitae::awesomecv | vitae::hyndman |
|---|---|
 |
 |
| vitae::latexcv | vitae::markdowncv |
|---|---|
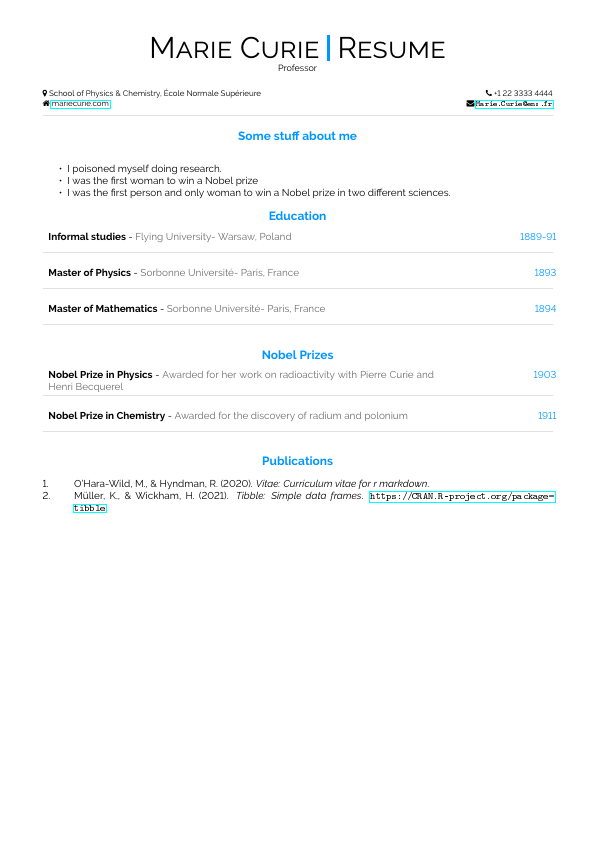 |
 |
| vitae::moderncv | vitae::twentyseconds |
|---|---|
 |
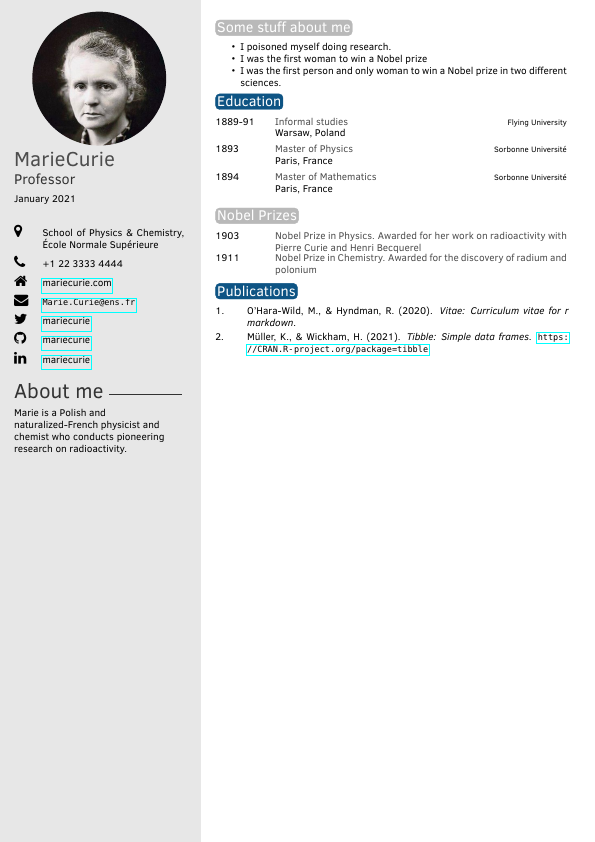 |
You can install the release version from CRAN.
install.packages('vitae')You can install the development version from GitHub.
# install.packages("devtools")
devtools::install_github("mitchelloharawild/vitae")This package requires LaTeX to be installed on your computer. If you’re encountering issues, please check that LaTeX is installed. The tinytex package makes it easy to setup LaTeX within R:
install.packages('tinytex')
tinytex::install_tinytex()The vitae package currently supports 6 popular CV templates, and adding more is a relatively simple process (details in the creating vitae templates vignette).
Creating a new CV with vitae can be done using the RStudio R Markdown
template selector: 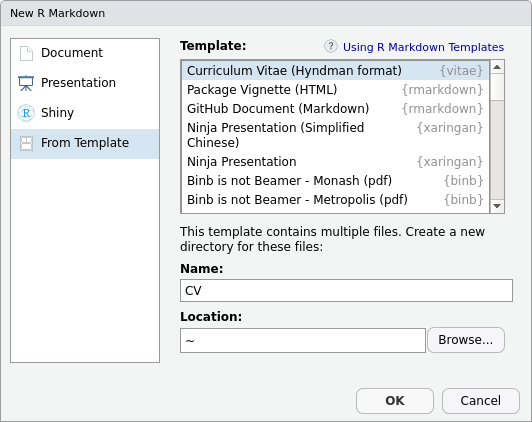
These templates leverage the strength of rmarkdown to include common
information in the YAML header (name, position, social links…) and
extended information in the main body. The main body of the CV is
written using markdown, and allows for data-driven generation of entries
using the *_entries
functions.
This allows you to import your working history from other sources (such
as ORCID, Google Scholar, or a maintained dataset), and include them
programmatically into your CV.
For example, the rorcid package can be used to extract Rob Hyndman’s education history:
orcid_data <- do.call("rbind",
rorcid::orcid_educations("0000-0002-2140-5352")$`0000-0002-2140-5352`$`affiliation-group`$summaries
)#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
#> education-summary.role-title education-summary.start-date.year.value
#> 1 PhD 1990
#> 2 Bachelor of Science (Honours) 1985
#> education-summary.end-date.year.value education-summary.organization.name
#> 1 1992 University of Melbourne
#> 2 1988 University of Melbourne
#> education-summary.organization.address.city
#> 1 Melbourne
#> 2 Melbourne
The package provides two types of entries from data, which are
detailed_entries and brief_entries. Both functions provide sections
for what, when, and with, and the detailed_entries additionally
supports where and why. These arguments support operations, so for
this example, we have used glue to combine the start and end years for
our when input. Excluding any inputs is also okay (as is done for
why), it will just be left blank in the CV.
orcid_data %>%
detailed_entries(
what = `education-summary.role-title`,
when = glue::glue("{`education-summary.start-date.year.value`} - {`education-summary.end-date.year.value`}"),
with = `education-summary.organization.name`,
where = `education-summary.organization.address.city`
)Additional examples of using this package can be found in the slides presented at ozunconf2018: https://slides.mitchelloharawild.com/vitae/
- Mitchell O’Hara-Wild
- Rob Hyndman
- Eric R. Scott
- Chris Umphlett
- Nat Price
- Sam Abbott (automatic deployment!)
- JooYoung Seo (printing multiple bibliographic entries according to a given csl file)
- Diogo M. Camacho
- Han Zhang (custom csl files)
- Bryan Jenks
- Lorena Abad
Add your vitae to the list using a PR.
Please note that the ‘vitae’ project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
The vitae project began as at rOpenSci’s OzUnconf 2018. A big thank you to rOpenSci and the event organisers for their work, which has played a big role in the formation of this package.


