ARMOR (Automated Reproducible MOdular RNA-seq) is a Snakemake workflow, aimed at performing a typical RNA-seq workflow in a reproducible, automated, and partially contained manner. It is implemented such that alternative or similar analysis can be added or removed.
ARMOR consists of a Snakefile, a conda environment file (envs/environment.yaml) a configuration file (config.yaml) and a set of R scripts, to perform quality control, preprocessing and differential expression analysis of RNA-seq data. The output can be combined with the iSEE R package to generate a shiny application for browsing and sharing the results.
By default, the pipeline performs all the steps shown in the diagram below. However, you can turn off any combination of the light-colored steps (e.g STAR alignment or DRIMSeq analysis) in the config.yaml file.
Advanced use: If you prefer other software to run one of the outlined steps (e.g. DESeq2 over edgeR, or kallisto over Salmon), you can use the software of your preference provided you have your own script(s), and change some lines within the Snakefile. If you think your "custom rule" might be of use to a broader audience, let us know by opening an issue.
Assuming that snakemake and conda are installed (and your system has the necessary libraries to compile R packages), you can use the following commands on a test dataset:
git clone https://github.com/csoneson/ARMOR.git
cd ARMOR && snakemake --use-conda
To use the ARMOR workflow on your own data, follow the steps outlined in the wiki.
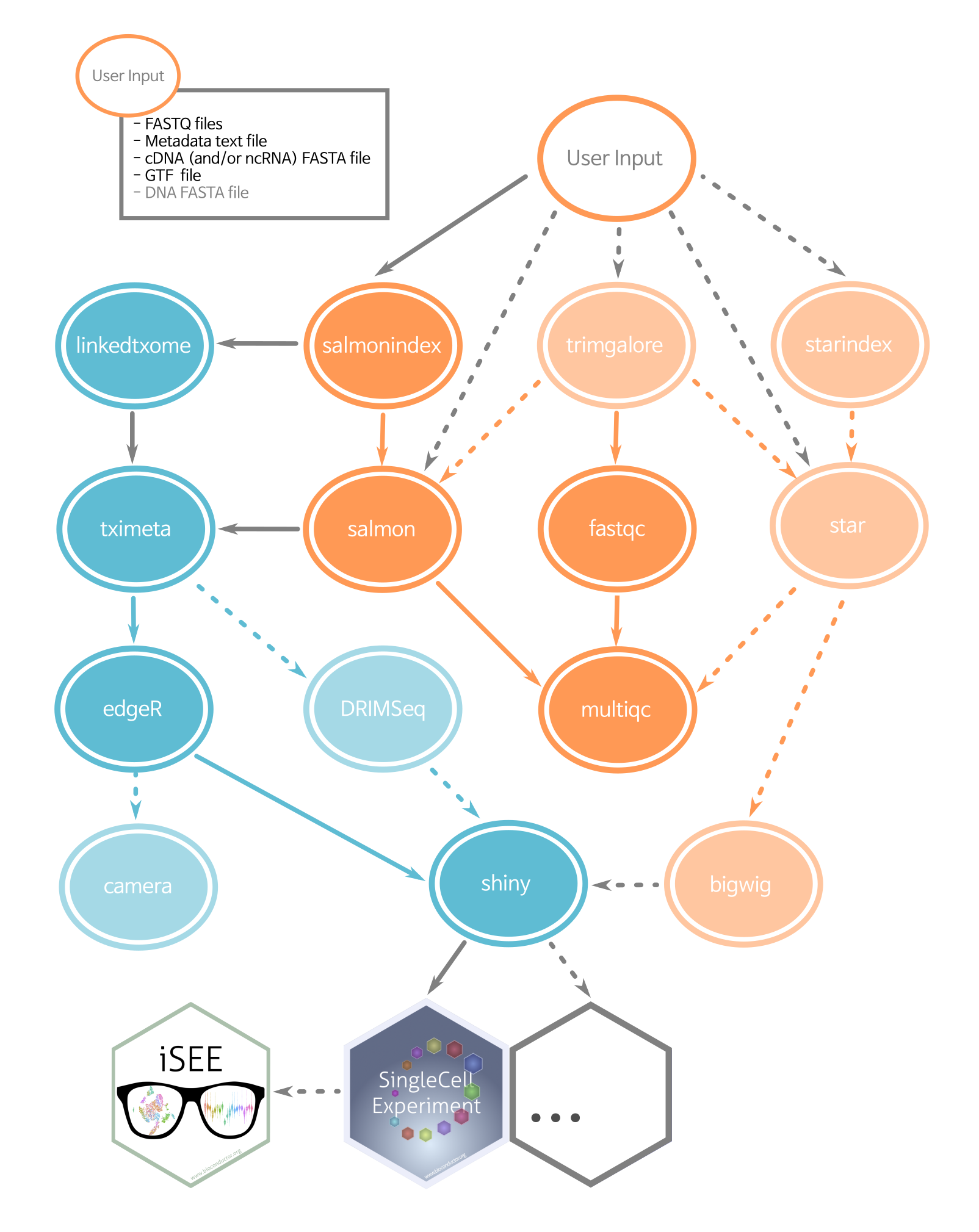
Blue circles are rules run in R, orange circles from software called as shell commands. Dashed lines and light-colored circles are optional rules, controlled in config.yaml
Current contributors include:
