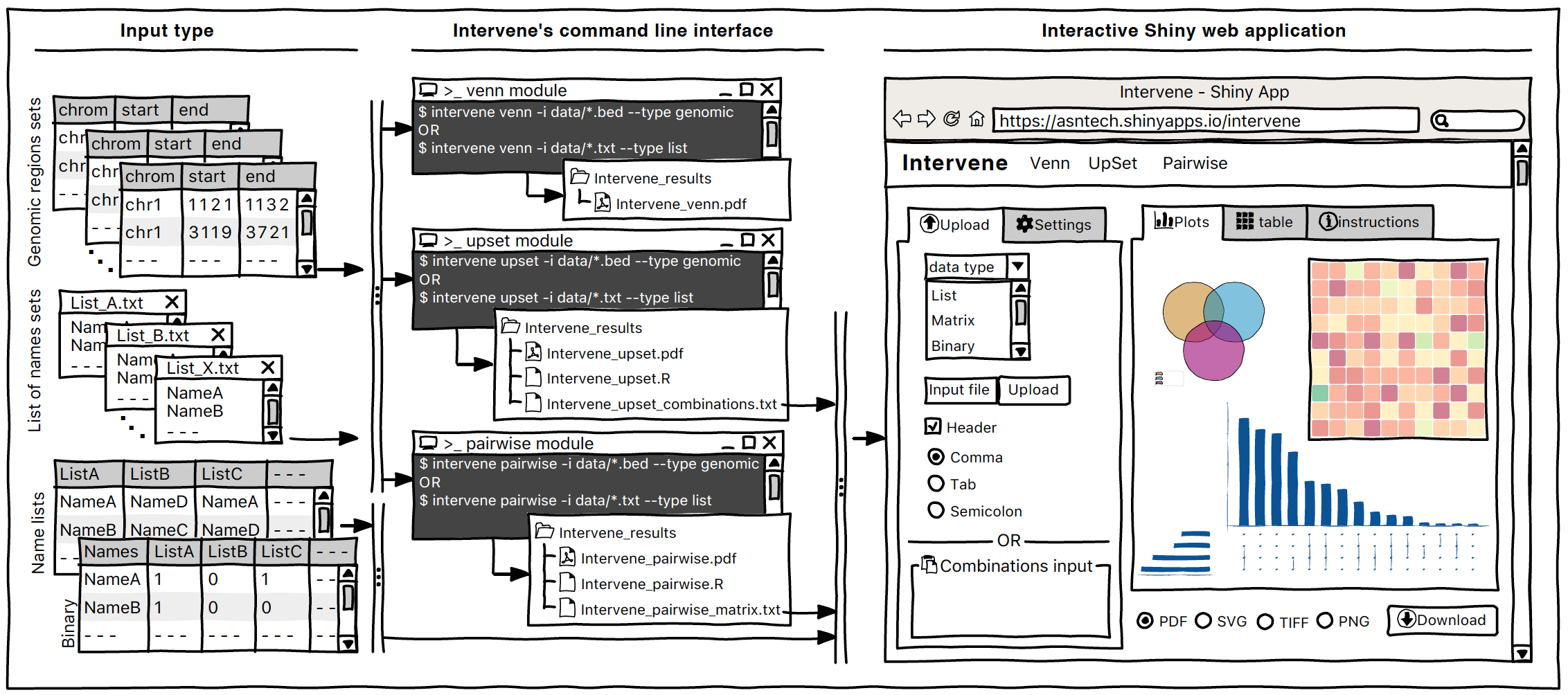
a tool for intersection and visualization of multiple gene or genomic region sets
A detailed documentation is available in different formats: HTML | PDF | ePUB
This will install all the dependencies and you are ready to use Intervene.
You can install Intervene from PyPi using pip.
Install from PyPi:
pip install interveneNote: If you install using pip, make sure to install BEDTools and R packages listed below.
Intervene requires the following Python modules and R packages:
- Python (=> 2.7 ): https://www.python.org/
- BedTools (Latest version): https://github.com/arq5x/bedtools2
- pybedtools (>= 0.7.9): https://daler.github.io/pybedtools/
- Pandas (>= 0.16.0): http://pandas.pydata.org/
- Seaborn (>= 0.7.1): http://seaborn.pydata.org/
- R (>= 3.0): https://www.r-project.org/
- R packages including UpSetR, corrplot
We are using pybedtools, which is Python wrapper for BEDTools. So, BEDTools should be installed before using Intervene. It's recomended to have a latest version, but if you have an older version already install, it should be fine.
A quick installation, if you have conda installed.
Please read the instructions at https://github.com/arq5x/bedtools2 to install BEDTools, and make sure it is on your path and you are able to call bedtools from any directory.
Intervene rquires three R packages, UpSetR , corrplot for visualization and Cairo to generate high-quality vector and bitmap figures.
You can install a development version by using git from GitHub or Bitbucket.
If you have git installed, use this:
If you have git installed, use this:
Once you have installed Intervene, you can type:
This will show the following help message.
usage: intervene <subcommand> [options]
positional arguments <subcommand>:
{venn,upset,pairwise}
List of subcommands
venn Venn diagram of intersection of genomic regions or list sets (upto 6-way).
upset UpSet diagram of intersection of genomic regions or list sets.
pairwise Pairwise intersection and heatmap of N genomic region sets in <BED/GTF/GFF> format.
optional arguments:
-h, --help show this help message and exit
-v, --version show program's version number and exitto see the help for the three subcommands pairwise, venn and upset type:
To run Intervene using example data, use the following commands. To access the test data make sure you have sudo or root access.
If you have installed Intervene locally from the source code, you may have problem to find test data. You can download the test data here https://github.com/asntech/intervene/tree/master/intervene/example_data and point to it using -i instead of --test.
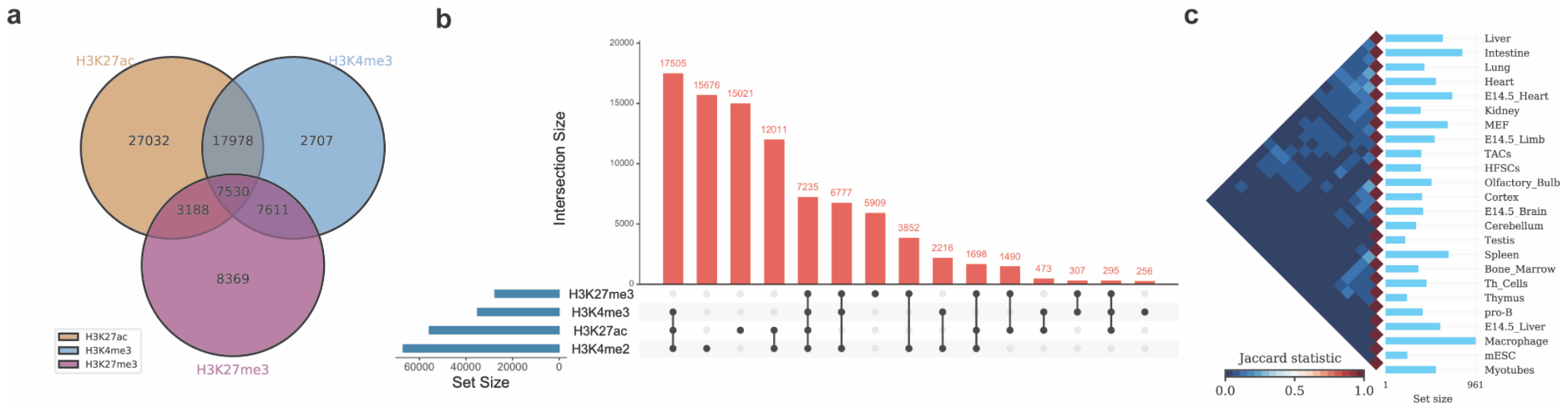
The above three test commands will generate the following three figures (a, b and c).
By default your results will stored in the current working directory with a folder named Intervene_results. If you wish to save the results in a specific folder, you can type:
intervene upset --test --output ~/path/to/your/folderIntervene Shiny App is freely available at https://asntech.shinyapps.io/intervene or https://intervene.shinyapps.io/intervene
The source code for the Shiny app is available at https://github.com/asntech/intervene-shiny
If you have questions, or found any bug in the program, please write to us at aziz.khan[at]ncmm.uio.no
If you use Intervene please cite us: Khan A, Mathelier A. Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinformatics. 2017;18:287. doi: 10.1186/s12859-017-1708-7