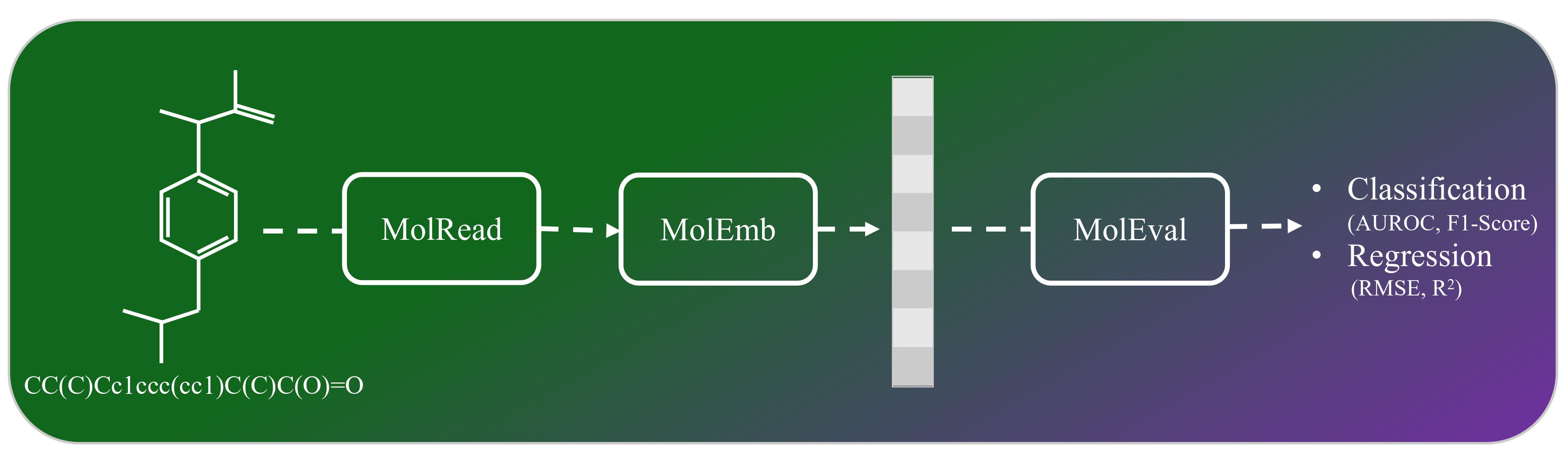
Drawing on the precedents set by SentEval—a toolkit designed to assess sentence embeddings— and MoleculeNet, a benchmark suite for molecular machine learning, we introduce MolEval. MolEval innovatively tackles the issue of evaluating large language models (LLMs) embeddings, which are traditionally expensive to execute on standard computing hardware. It achieves this by offering a repository of pre-computed molecule embeddings, alongside a versatile platform that facilitates the evaluation of any embeddings derived from molecular structures. This approach not only streamlines the assessment process but also makes it more accessible to researchers and practitioners in the field.
The following are the features in this toolkit:
!git clone https://github.com/sshaghayeghs/MolEval
!cd MolEval
!pip install torch transformers pandas numpy tqdm openai deepchem rdkit networkx matplotlibAvailable datasets from MoleculeNet: bbbp, bace_classifcation, hiv, tox21, clintox, sider, lipo, freesolv, delaney
from MolEval.MolRead import load_dataset
df=load_dataset('bace_classification')from MolEval.MolGraph import MolGraph
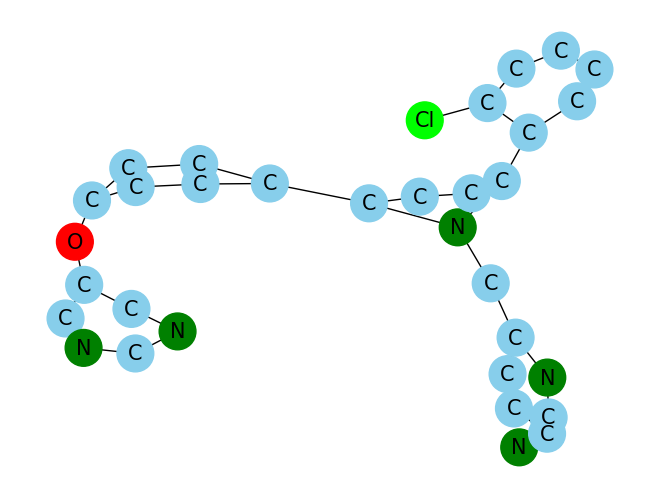
print(df['SMILES'][100])
MolGraph(df['SMILES'][100])Clc1ccccc1-c1n(Cc2nc(N)ccc2)c(cc1)-c1ccc(Oc2cncnc2)cc1
Available embedding model: SBERT, LLaMA2, Molformer, ChemBERTa, BERT, RoBERTa_ZINC, RoBERTa, SimCSE, AngleBERT, GPT, Mol2Vec, Morgan
from MolEval import MolEmb
model_name = 'Morgan' # Replace with the model you want to use
openai_api_key = 'your_openai_api_key' # Required if using GPT
huggingface_token = 'your_huggingface_token' # Required if using LLaMA2
extractor = MolEmb.EmbeddingExtractor(model_name=model_name, df=df, openai_api_key=openai_api_key, huggingface_token=huggingface_token)
emb, df = extractor.get_embeddings()
print(emb)If dataset in bbbp, bace_classification, hiv, task is Classification
elif dataset in tox21, clintox, sider, task is MultitaskClassification
from MolEval.MolEval import evaluate_classification
f1_score,f1_score_std,AUROC,AUROC_std=evaluate_classification(features=emb.to_numpy(), targets=df.drop(columns=['SMILES']).to_numpy(), n_splits=5, task='Classification')
print(f'F1 score: {f1_score:.4f} +/- {f1_score_std:.4f}')
print(f'AUROC: {AUROC:.4f} +/- {AUROC_std:.4f}')If dataset in lipo, freesolv, delaney, use is evaluate_regression
from MolEval import evaluate_regression
RMSE,RMSE_std,R2,R2_std=evaluate_regression(features=emb.to_numpy(), targets=df.drop(columns=['SMILES']).to_numpy(), n_splits=5)
print(f'RMSE: {RMSE:.4f} +/- {RMSE_std:.4f}')
print(f'R2: {R2:.4f} +/- {R2_std:.4f}')@inproceedings{sadeghi2024moleval,
title={MolEval: An Evaluation Toolkit for Molecular Embeddings via LLMs},
author={Sadeghi, Shaghayegh and Forooghi, Ali and Lu, Jianguo and Ngom, Alioune},
booktitle={ICML 2024 Workshop on Efficient and Accessible Foundation Models for Biological Discovery}
}