We are proud to introduce Asclepius, a more advanced clinical large language model. As this model was trained on synthetic clinical notes, it is publicly accessible via Huggingface. If you are considering using CAMEL, we highly recommend switching to Asclepius instead. For more information, please visit this link.
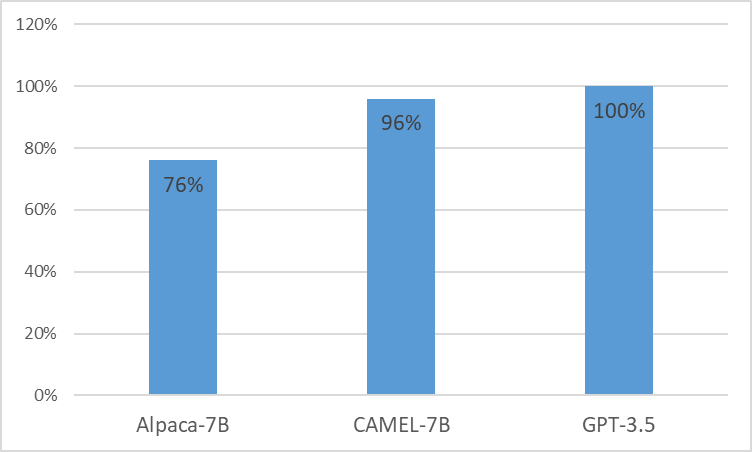
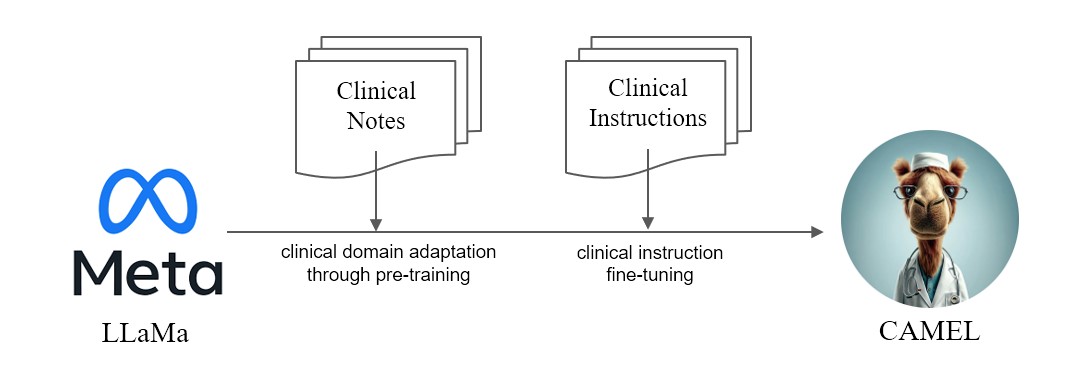
We present CAMEL, Clinically Adapted Model Enhanced from LLaMA. As LLaMA for its foundation, CAMEL is furtherpre-trained on MIMIC-III and MIMIC-IV clinical notes, and finetuned over clinical instructions (Figure 2). Our preliminary evaluation with GPT-4 assessment, demonstrates that CAMEL achieves over 96% of the quality of OpenAI's GPT-3.5 (Figure 1). In accordance with the data usage policies of our source data, both our instruction dataset and model will be published on PhysioNet with credentialized access. To facilitate replication, we will also release all code, allowing individual healthcare institutions to reproduce our model using their own clinical notes. For further detail, please refer our blog post.
Due to the license issue of MIMIC and i2b2 datasets, we cannot publish the instruction dataset and checkpoints. We would publish our model and data via physionet within few weeks.
Environment Setup
conda create -n camel python=3.9 -y
conda activate camel
conda install pytorch torchvision torchaudio pytorch-cuda=11.8 -c pytorch -c nvidia -y
pip install pandarallel pandas jupyter numpy datasets sentencepiece openai fire
pip install git+https://github.com/huggingface/transformers.git@871598be552c38537bc047a409b4a6840ba1c1e4
Pretraining
- Note Preprocessing
- For each note, we concatenate the category in front of the text.
- To prevent test set leakage, we removed 404 overlapping notes from MIMIC-III with RadQA, CLIP, n2c2 2018 datasets for further evaluation.
- We concatenate all notes with
<eos>tokens. $ python pretraining_preprocess/mimiciii_preproc.py --mimiciii_note_path {MIMICIII_NOTE_PATH} --output_path {OUTPUT_PATH}$ python pretraining_preprocess/mimiciv_preproc.py --discharge_note_path {DISCHAGE_NOTE_PATH} --radiology_note_path {RADIOLOGY_NOTE_PATH} --output_path {OUTPUT_PATH}$ python pretraining_preprocess/tokenize_data.py --data_path {DATA_PATH} --save_path {SAVE_PATH}
- Run Pretriaining
$ torchrun --nproc_per_node=8 --master_port={YOUR_PORT} \ src/train.py \ --model_name_or_path "decapoda-research/llama-7b-hf" \ --data_path {DATA_FILE} \ --bf16 True \ --output_dir ./checkpoints \ --num_train_epochs 1 \ --per_device_train_batch_size 2 \ --per_device_eval_batch_size 2 \ --gradient_accumulation_steps 8 \ --evaluation_strategy "no" \ --save_strategy "steps" \ --save_steps 1000 \ --learning_rate 2e-5 \ --weight_decay 0. \ --warmup_ratio 0.03 \ --lr_scheduler_type "cosine" \ --logging_steps 1 \ --fsdp "full_shard auto_wrap" \ --fsdp_transformer_layer_cls_to_wrap 'LlamaDecoderLayer' \ --tf32 True \ --model_max_length 2048 \ --gradient_checkpointing True
Instruction Finetuning
-
NOTE: To generate instructions, you should use certified Azure Openai API.
-
Instruction Generation
- Set environment variables
OPENAI_API_KEYOPENAI_API_BASEOPENAI_DEPLOYMENT_NAME
- Preprocess Notes
$ python instructino/preprocess_note.py
- De-Identification instruction generation
$ python instruction/de_id_gen.py --input {PREPROCESSED_NOTES} --output {OUTPUT_FILE_1} --mode inst$ python instruction/de_id_postprocess.py --input {OUTPUT_FILE_1} --output {OUTPUT_FILE_2}$ python instruction/de_id_gen.py --input {OUTPUT__FILE_2} --output {inst_output/OUTPUT_FILE_deid} --mode ans
- Other tasks instruction generation
- You can generate instructions selectively for each dataset.
$ python instruction/instructtion_gen.py --input {PREPROCESSED_NOTES} --output {inst_output/OUTPUT_FILE} --source {mimiciii, mimiciv, i2b2}
- Merge and formatting files
$ python instruction/merge_data.py --data_path {inst_output} --output {OUTPUT_FILE_FINAL}
- Set environment variables
-
Run Instruction Finetuning
- All of our experimente were perfomed with 8x A6000 gpus.
- Adjust
nproc_per_nodeandgradient accumulate stepto fit to your hardware (global batch size=128).
$ torchrun --nproc_per_node=8 --master_port={YOUR_PORT} \
src/instruction_ft.py \
--model_name_or_path "decapoda-research/llama-7b-hf" \
--data_path {OUTPUT_FILE_FINAL} \
--bf16 True \
--output_dir ./checkpoints \
--num_train_epochs 3 \
--per_device_train_batch_size 2 \
--per_device_eval_batch_size 2 \
--gradient_accumulation_steps 8 \
--evaluation_strategy "no" \
--save_strategy "epoch" \
--learning_rate 2e-5 \
--weight_decay 0. \
--warmup_ratio 0.03 \
--lr_scheduler_type "cosine" \
--logging_steps 1 \
--fsdp "full_shard auto_wrap" \
--fsdp_transformer_layer_cls_to_wrap 'LlamaDecoderLayer' \
--tf32 True \
--model_max_length 2048 \
--gradient_checkpointing True
--ddp_timeout 18000
Evaluation
-
Run model on MTSamples
CUDA_VISIBLE_DEVICES=0 python src/evaluate.py \ --model_name {MODEL_PATH} \ --data_path eval/mtsamples_instructions.json \ --output_path {OUTPUT_PATH}- We provide the output generated by GPT-3.5, Alpaca, and CAMEL as
mtsamples_results.jsoninevalfolder.
- We provide the output generated by GPT-3.5, Alpaca, and CAMEL as
-
Run GPT-4 for evaluation
python eval/gpt4_evaluate.py --input {INPUT_PATH} --output {OUTPUT_PATH}
@misc{CAMEL,
title = {CAMEL : Clinically Adapted Model Enhanced from LLaMA},
author = {Sunjun Kweon and Junu Kim and Seongsu Bae and Eunbyeol Cho and Sujeong Im and Jiyoun Kim and Gyubok Lee and JongHak Moon and JeongWoo Oh and Edward Choi},
month = {May},
year = {2023}
publisher = {GitHub},
journal = {GitHub repository},
howpublished = {\url{https://github.com/starmpcc/CAMEL}},
}