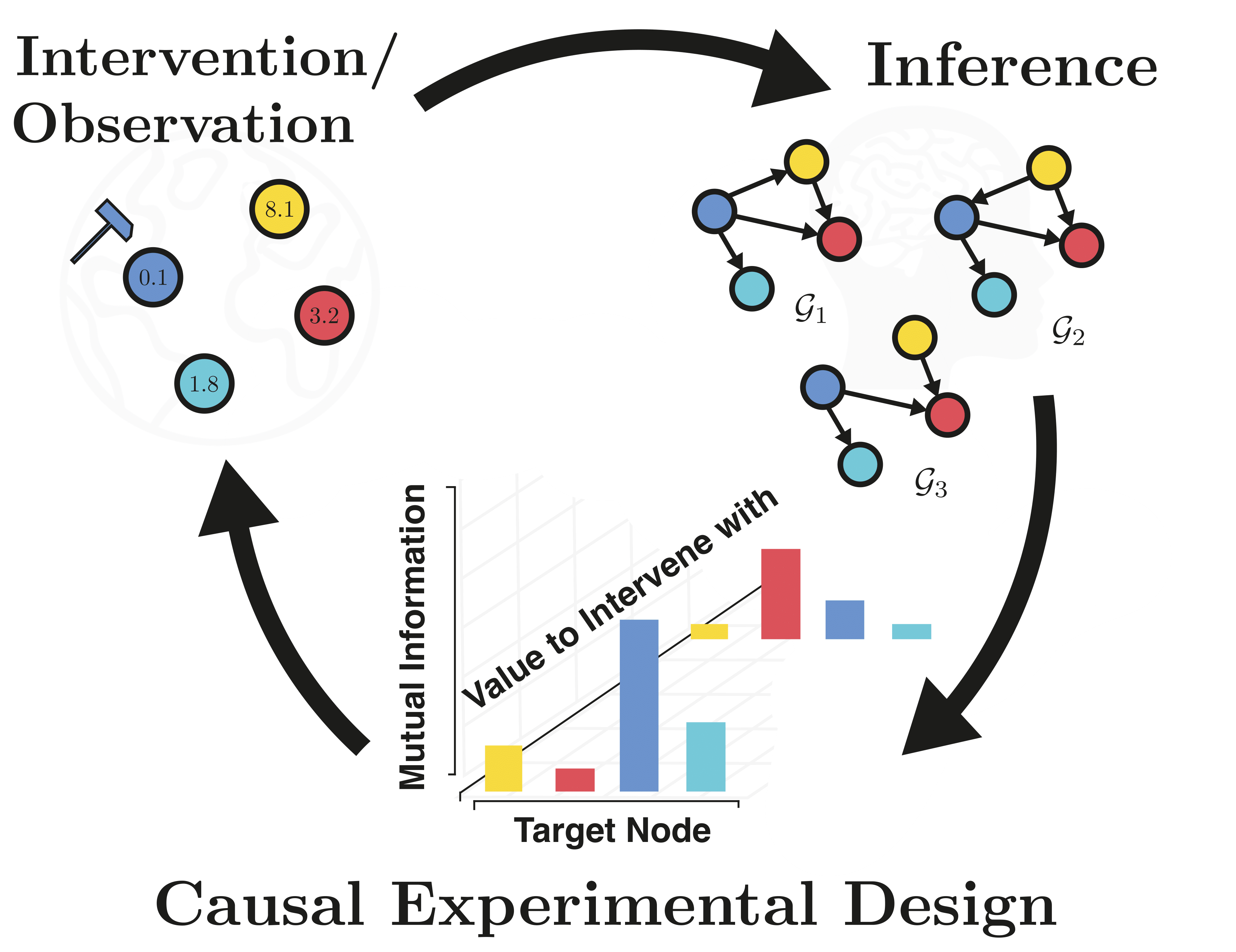
This is the official repository with the python implementation for the paper "Interventions, Where and How? Experimental Design for Causal Models at Scale", NeurIPS (2022).
The repository contains different acquisition strategies for optimal targeted interventions to learn causal models, including the proposed approach CBED (which acquires both intervention target and value). The provided repository further contains two different batching strategies: the greedy strategy and the stochastic batch strategy.
In order to run this repository on your machine, it is recommended to follow the below steps.
- Clone the repository
git clone --recurse-submodules https://github.com/yannadani/cbed.git
cd cbed- Install the relevant packages in an environment, either through pip or Anaconda. Anaconda is preferable as R packages can be installed easily without much effort. To do so, please do the following:
conda env create -f environment.yml
conda activate cbed_envIt is also possible to run the proposed CBED with just a venv. However, you will not be able compute the Structural Interventional Distance (SID) which requires R. In addition, R is also required by DAG bootstrap. If you are not planninng to use R (or have R already installed), run the following:
python3 -m venv cbed_env
source cbed_env/bin/activate- Install the requirements by running the following commands:
pip install -r requirements.txt
pip install --upgrade "jax[cuda]" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html
pip install -e models/dibsIf using R, then run:
Rscript models/dag_bootstrap_lib/install.rThe posterior models to estimate the expected information gain are located inside models. These include DiBS and DAG Bootstrap with GIES. In the paper, we use DAG bootstrap for the linear models and DiBS for nonlinear models.
- In order to run the CBED acquisition strategy for nonlinear SCM for 50 nodes, run the following command:
python experimental_design.py --nonlinear --model dibs_nonlinear --num_nodes 50 --strategy softcbed --value_strategy gp-ucb - If
Ris not installed, run the following command:
python experimental_design.py --nonlinear --model dibs_nonlinear --num_nodes 50 --strategy softcbed --value_strategy gp-ucb --no_sid- For running on the DREAM environment, run the following command:
python experimental_design.py --num_nodes 50 --model dibs_nonlinear --noise_type gaussian --strategy softcbed --env dream4 --dream4_path envs/dream4/configurations --dream4_name InSilicoSize50-Yeast1- To reproduce the results in the paper, run the following command for nonlinear SCM:
bash run_experiments.sh --nonlinear --model dibs_nonlinear --strategy <strategy> --value_strategy <value_strategy> --num_nodes <num_nodes>where strategy is one of cbed, greedycbed, softcbed, random, ait and value_strategy is one of gp-ucb, fixed, sample-dist. Num nodes is any number of nodes of your choice. In the paper, we run it for 50 and 20 nodes.
- For running linear SCMs, run the following command:
bash run_experiments.sh --model dag_bootstrap --strategy <strategy> --value_strategy <value_strategy> --num_nodes <num_nodes>with the arguments the same as before.
Note: If you are running experiments of different strategies with same data seeds on different machines, JAX can show non-deterministic behaviour which might mean the initial model after having trained on observational data could be differernt. In order to counteract this, set the environment variable XLA_FLAGS='--xla_gpu_deterministic_reductions --xla_gpu_autotune_level=0'.
- Finally, to reproduce the entire set of experiments on DREAM, run the following command and follow the ensuing commands shown on the screen:
wandb sweep dream_sweep.ymlThis code is official implementation of the following paper:
Panagiotis Tigas, Yashas Annadani, Andrew Jesson, Bernhard Schölkopf, Yarin Gal and Stefan Bauer. Interventions, Where and How? Experimental Design for Causal Models at Scale. In Advances of Neural Information Processing Systems (NeurIPS), 2022. PDF
If this code was useful, please consider citing this work.
@article{tigas2022interventions,
title={Interventions, where and how? experimental design for causal models at scale},
author={Tigas, Panagiotis and Annadani, Yashas and Jesson, Andrew and Sch{\"o}lkopf, Bernhard and Gal, Yarin and Bauer, Stefan},
journal={Advances in Neural Information Processing Systems},
year={2022}
}