Version 0.1
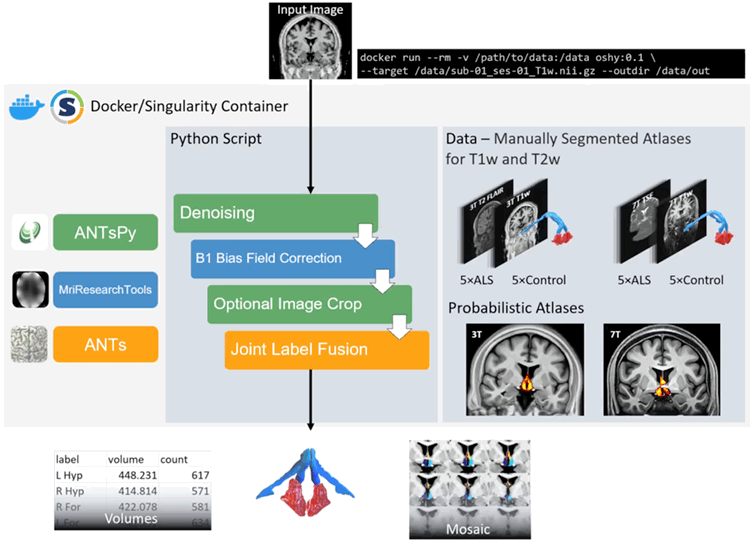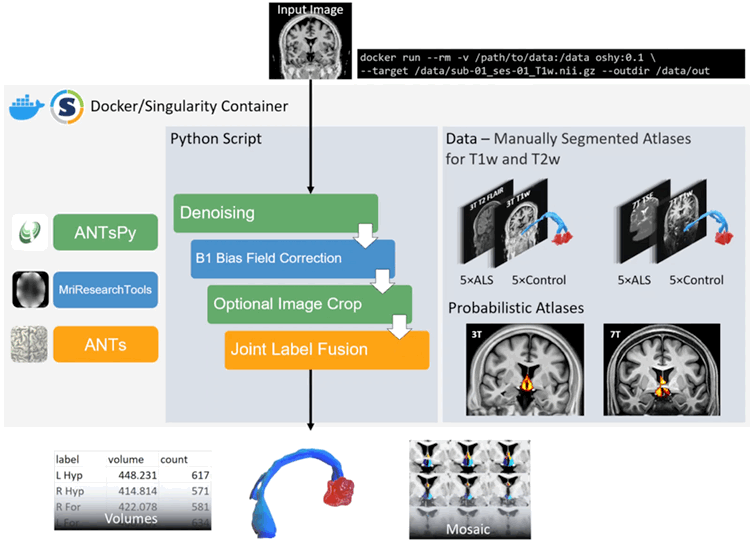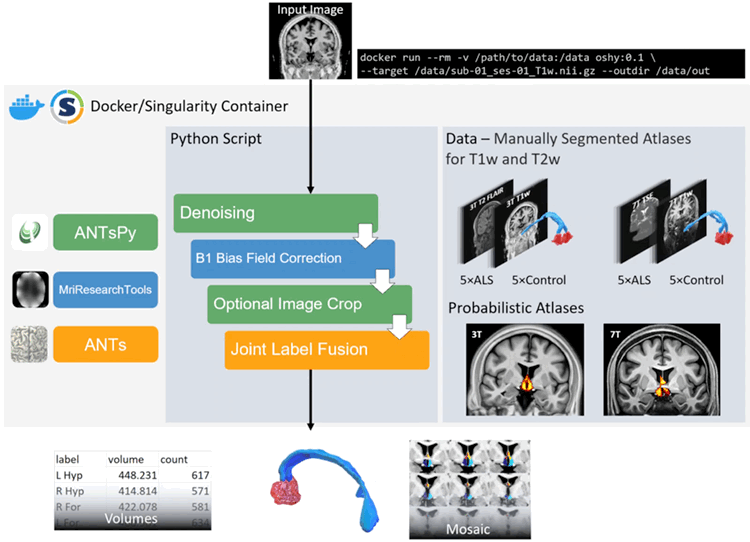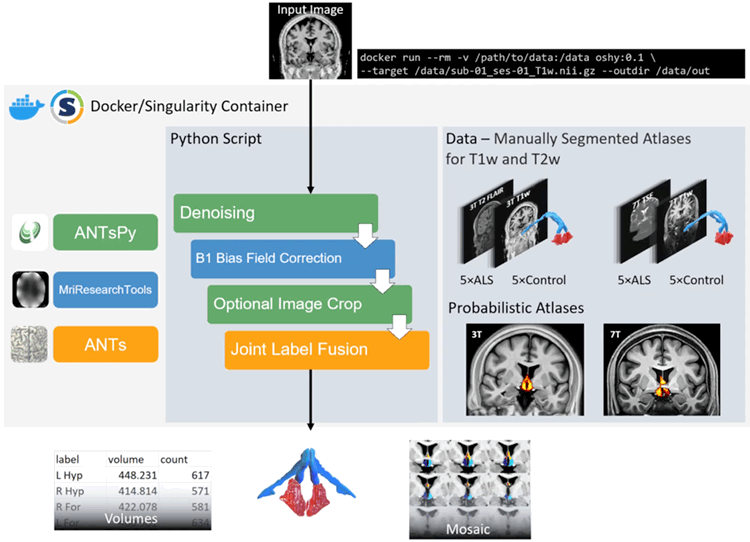
OSHy-X is an atlas repository (https://osf.io/zge9t) and containerised Python script that automatically segments the hypothalamus and fornix at 3T and 7T in both T1w and T2w scans.
If you intend to use OSHy-X, please cite this abstract: TBA
You have the option of running OSHy-X via NeuroDesk, a Docker container by itself, or a Singularity container.
Follow instructions here to install Neurodesk.
- Install Docker here.
- Open a terminal and run:
docker pull jerync/oshyx_0.1:20211126
To pull the container. Or run:
docker run --rm -v /path/to/data/folder/:/data/ jerync/oshyx_0.1:20211126 --target /data/input_file.nii.gz --outdir /data/output_directory
- Install Singularity here.
- Open a terminal and run
singularity pull docker://jerync/oshyx_0.1:20211126 oshyx_0.1.sifTo pull the container.
Usage: docker run --rm -v /path/to/data:/data jerync/oshyx_0.1:20211126
[-h] -t TARGET [TARGET ...] -o OUTDIR [-c CROP] [-w WEIGHTING]
[-d DENOISE] [-f FIELDCORRECTION] [-m MOSAIC] [-x TESLA]
[-b BIMODAL] [-n NTHREADS]
singularity run -B /path/to/data:/data oshyx_0.1.sif
[-h] -t TARGET [TARGET ...] -o OUTDIR [-c CROP] [-w WEIGHTING]
[-d DENOISE] [-f FIELDCORRECTION] [-m MOSAIC] [-x TESLA]
[-b BIMODAL] [-n NTHREADS]
Options:
-h, --help Show this help message and exit
-t TARGET [TARGET ...], --target TARGET [TARGET ...]
A string or list of strings pointing to the target
image(s). Must be a NIfTI file. For a test run,
specify /OSHy/sub-test.nii.gz
-o OUTDIR, --outdir OUTDIR
A string pointing to the output directory. Please
ensure this is within the mounted volume (Specified
with the -v flag for the docker run command.
-c CROP, --crop CROP Optional. A boolean indicating if the target image and
priors are to be cropped. If False, whole-image priors
will be used, which will improve the segmentation but
significantly increase the runtime. (default: True)
-w WEIGHTING, --weighting WEIGHTING
A string indicating the weighting of the input
image(s). This can be either T1w or T2w. (default: T1w
-d DENOISE, --denoise DENOISE
Optional. A boolean indicating if denoising is to be
run on the target image. (default: True)
-f FIELDCORRECTION, --fieldCorrection FIELDCORRECTION
Optional. A boolean indicating if B1 bias field
correction is to be run on the target image. (default:
True)
-m MOSAIC, --mosaic MOSAIC
Optional. A boolean indicating if a mosaic image is to
be plotted after running Joint Label Fusion. (default:
True)
-x TESLA, --tesla TESLA
Optional. An integer (either 3 or 7) indicating the
field strength. (default: 3)
-b BIMODAL, --bimodal BIMODAL
Optional. A boolean indicating if bimodal priors are
to be used. If FALSE, then only unimodal priors
(specified in --weighting) will be used.(default:
False)
-n NTHREADS, --nthreads NTHREADS
Optional. An integer indicating the number of threads.
This is passed to the global variable
ITK_GLOBAL_DEFAULT_NUMBER_OF_THREADS and the -j flag
in Joint Label Fusion. (default: 6)
All output is written to the output directory (specified using the -o/--outdir flag.)
Contents of the output include:
sub-XX_Labels.nii.gz: Output from Joint Label Fusion. The label file for the left and right hemispheres of the Hypothalamus and Fornix. If--cropisTruethen this label file will also be cropped. The labels are as follows:- Left Hypothalamus
- Right Hypothalamus
- Right Fornix
- Left Fornix
sub-XX_ressampled_Labels.nii.gz: sub-XX_Labels.nii.gz but resampled to the input target image.sub-XX_hypothalamus.nii.gz: sub-XX_resampled_Labels.nii.gz but with only hypothalamus labels.sub-XX_fornix.nii.gz: sub-XX_resampled_Labels.nii.gz but with only fornix labels.sub-XX_mosaic.png: A 16 slice coronal visualisation of the segmentation.sub-XX_volumes.csv: Volumes of the four labels (as described above). Units for volume are in mm3.