Author: Samuel Farrens
Year: 2018
Version: 5.0
Email: samuel.farrens@gmail.com
Website: sfarrens.github.io
Reference Paper: arXiv:1703.02305
This repository contains a Python code designed for PSF deconvolution and analysis.
The directory lib contains several primary functions and classes, but the majority of the optimisation and analysis tools are provided in sf_tools.
After downloading or cloning the repository simply run:
$ python setup.py installIn order to run sf_deconvolve the following packages must be installed:
-
Python [Tested with v 2.7.11 and 3.6.3]
-
ModOpt [>=1.1.4]
-
sf_tools [>=2.0]
-
The current implementation of wavelet transformations additionally requires the
mr_transform.ccC++ script from the Sparse2D library (https://github.com/CosmoStat/Sparse2D). These C++ scripts will be need to be compiled in order to run. Note: The low-rank approximation method can be run purely in Python without the Sparse2D binaries.
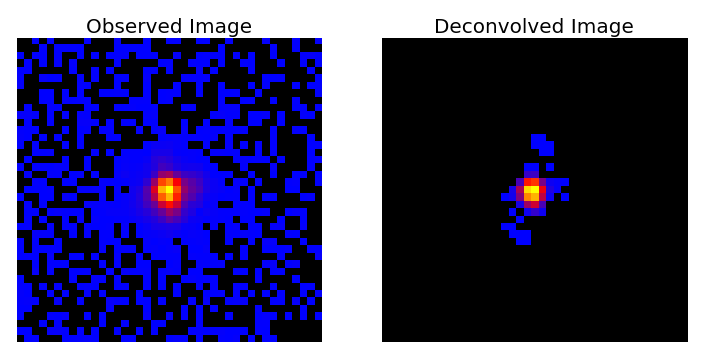
The primary code is an executable script called sf_deconvolve.py which is designed to take an observed (i.e. with PSF effects and noise) stack of galaxy images and a known PSF, and attempt to reconstruct the original images. The input format are Numpy binary files (.npy) or FITS image files (.fits).
The input files should have the following format:
-
Input Images: This should be either a Numpy binary or a FITS file containing a 3D array of galaxy images. e.g. for a sample of 10 images, each with size 41x41, the shape of the array should be [10, 41, 41].
-
Input PSF(s): This should be either a Numpy binary or a FITS file containing a 2D array (for a fixed PSF) or a 3D array (for a spatially varying PSF) of PSF images. For the spatially varying case the number of PSF images must match the number of corresponding galaxy images. e.g. For a sample of 10 images the codes expects 10 PSFs.
See the files provided in the examples directory for reference.
The code can be run in a terminal (not in a Python session) as follows:
$ sf_deconvolve.py -i INPUT_IMAGES.npy -p PSF.npy -o OUTPUT_NAMEWhere INPUT_IMAGES.npy denotes the Numpy binary file containing the stack of observed galaxy images, PSF.npy denotes the PSF corresponding to each galaxy image and OUTPUT_NAME specifies the output path and file name.
Alternatively the code arguments can be stored in a configuration file (with any name) and the code can be run by providing
the file name preceded by a @.
$ sf_deconvolve.py @config.iniAn example configuration file is provided in the examples directory.
The code can be run in an active Python session in two ways. For either approach first import sf_deconvolve:
>>> import sf_deconvolveThe first approach simply runs the full script where the command line arguments can be passed as a list of strings:
>>> sf_deconvolve.main(['-i', 'INPUT_IMAGES.npy', '-p', 'PSF.npy', '-o', 'OUTPUT_NAME'])The second approach assumes that the user has already has read the images and PSF(s) into memory and wishes to return the deconvolution results to memory:
>>> opts = vars(sf_deconvolve.get_opts(['-i', 'INPUT_IMAGES.npy', '-p', 'PSF.npy', '-o', 'OUTPUT_NAME']))
>>> primal_res, dual_res, psf_res = sf_deconvolve.run(INPUT_IMAGES, INPUT_PSFS, **opts)Where INPUT_IMAGES and INPUT_PSFS are both Numpy arrays. The resulting deconvolved images will be saved to the variable primal_res.
In both cases it is possible to read a predefined configuration file.
>>> opts = vars(sf_deconvolve.get_opts(['@config.ini']))
>>> primal_res, dual_res, psf_res = sf_deconvolve.run(INPUT_IMAGES, INPUT_PSFS, **opts)The following example can be run on the sample data provided in the example directory.
This example takes a sample of 100 galaxy images (with PSF effects and added noise) and the corresponding PSFs, and recovers the original images using low-rank approximation via Condat-Vu optimisation.
$ sf_deconvolve.py -i example_image_stack.npy -p example_psfs.npy -o example_output --mode lowrThe example can also be run using the configuration file provided.
The result will be two Numpy binary files called example_output_primal.npy and example_output_dual.npy corresponding to the primal and dual variables in the splitting algorithm. The reconstructed images will be in the example_output_primal.npy file.
The example can also be run with the FITS files provided.
-
-i INPUT, --input INPUT: Input data file name. File should be a Numpy binary containing a stack of noisy galaxy images with PSF effects (i.e. a 3D array).
-
-p PSF, --psf PSF: PSF file name. File should be a Numpy binary containing either: (a) a single PSF (i.e. a 2D array for fixed format) or (b) a stack of PSFs corresponding to each of the galaxy images (i.e. a 3D array for obj_var format).
-
-h, --help: Show the help message and exit.
-
-v, --version: Show the program's version number and exit.
-
-q, --quiet: Suppress verbose for each iteration.
-
-o, --output: Output file name. If not specified output files will placed in input file path.
-
--output_format Output file format [npy or fits].
Initialisation:
-
-k, --current_res: Current deconvolution results file name (i.e. the file containing the primal results from a previous run).
-
--noise_est: Initial estimate of the noise standard deviation in the observed galaxy images. If not specified this quantity is automatically calculated using the median absolute deviation of the input image(s).
Optimisation:
-
-m, --mode {all,sparse,lowr,grad}: Option to specify the optimisation mode [all, sparse, lowr or grad]. all performs optimisation using both low-rank approximation and sparsity, sparse using only sparsity, lowr uses only low-rank and grad uses only gradient descent. (default: lowr)
-
--opt_type {condat,fwbw,gfwbw}: Option to specify the optimisation method to be implemented [condat, fwbw or gfwbw]. condat implements the Condat-Vu proximal splitting method, fwbw implements Forward-Backward splitting with FISTA speed-up and gfwbw implements the generalised Forward-Backward splitting method. (default: condat)
-
--n_iter: Number of iterations. (default: 150)
-
--cost_window: Window to measure cost function (i.e. interval of iterations for which cost should be calculated). (default: 1)
-
--convergence: Convergence tolerance. (default: 0.0001)
-
--no_pos: Option to turn off positivity constraint.
-
--grad_type: Option to specify the type of gradient [psf_known, psf_unknown, none]. psf_known implements deconvolution with PSFs provided, psf_unknown simultaneously improves the PSF while performing the deconvolution, none implements deconvolution without gradient descent (for testing purposes only). (default: psf_known)
Low-Rank Aproximation:
-
--lowr_thresh_factor: Low rank threshold factor. (default: 1)
-
--lowr_type: Type of low-rank regularisation [standard or ngole]. (default: standard)
-
--lowr_thresh_type: Low rank threshold type [soft or hard]. (default: hard)
Sparsity:
-
--wavelet_type: Type of Wavelet to be used (see iSap Documentation). (default: 1)
-
--wave_thresh_factor: Wavelet threshold factor. (default: [3.0, 3.0, 4.0])
-
--n_reweights: Number of reweightings. (default: 1)
PSF Estimation
- --lambda_psf: Regularisation control parameter for PSF estimation. (default: 1.0)
- --beta_psf: Gradient step for PSF estimation. (default: 1.0)
Condat Algorithm:
-
--relax: Relaxation parameter (rho_n in Condat-Vu method). (default: 0.8)
-
--condat_sigma: Condat proximal dual parameter. If the option is provided without any value, an appropriate value is calculated automatically. (default: 0.5)
-
--condat_tau: Condat proximal primal parameter. If the option is provided without any value, an appropriate value is calculated automatically. (default: 0.5)
Testing:
-
-c, --clean_data: Clean data file name.
-
-r, --random_seed: Random seed. Use this option if the input data is a randomly selected subset (with known seed) of the full sample of clean data.
-
--true_psf: True PSFs file name.
-
--kernel: Standard deviation of pixels for Gaussian kernel. This option will multiply the deconvolution results by a Gaussian kernel.
-
--metric: Metric to average errors [median or mean]. (default: median)
- If you get the following error:
ERROR: svd() got an unexpected keyword argument 'lapack_driver'
Update your Numpy and Scipy installations
$ pip install --upgrade numpy
$ pip install --upgrade scipy