Please click to watch the overview video
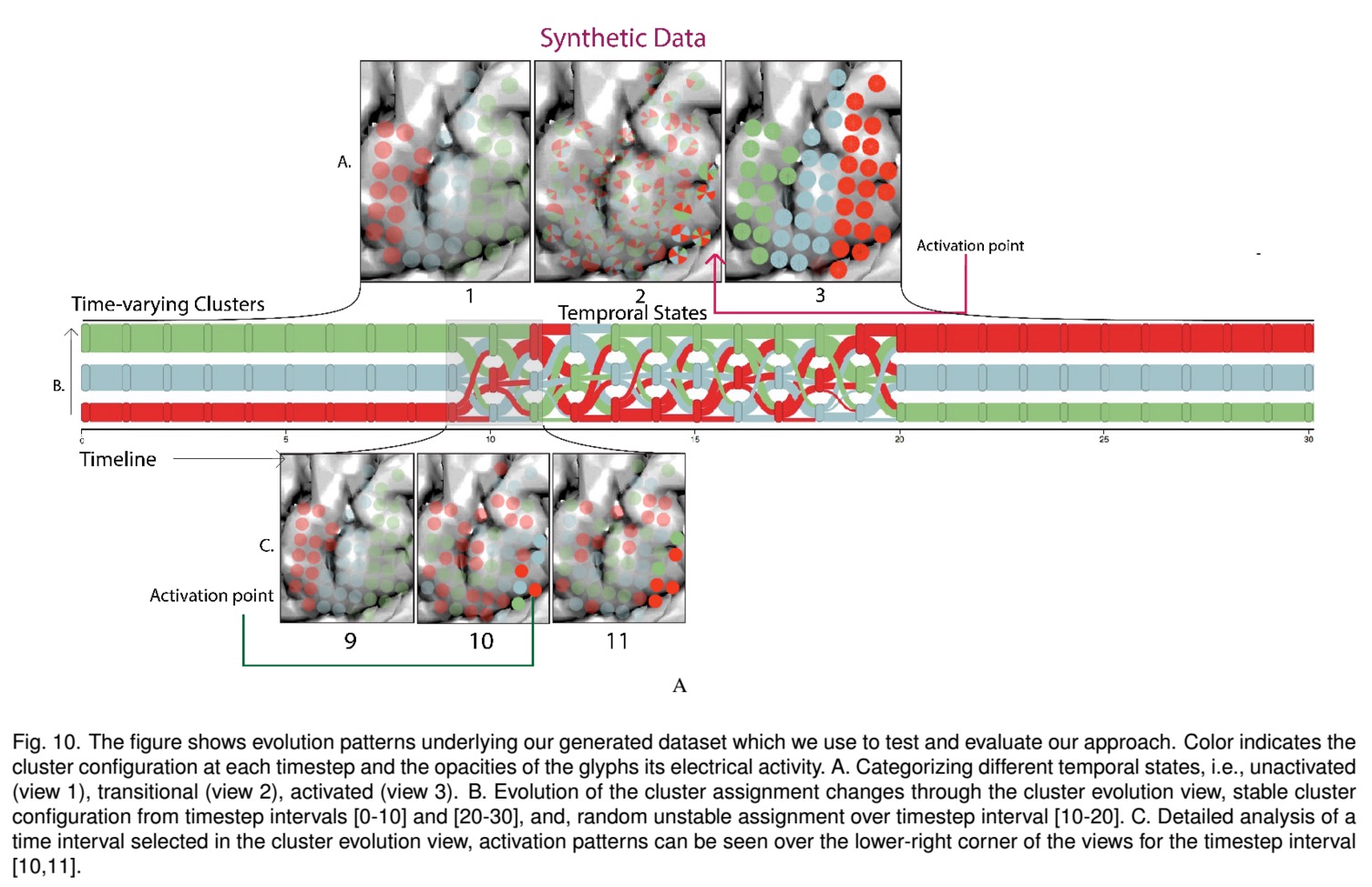
We present ECoG ClusterFlow, an interactive visual analysis system for the exploration of high-resolution Electrocorticography (ECoG) data. Our system detects and visualizes dynamic high-level structures, such as communities, using the time-varying spatial connectivity network derived from the high-resolution ECoG data. ECoG ClusterFlow provides a multi-scale visualization of the spatio-temporal patterns underlying the time-varying communities using two views: 1) an overview summarizing the evolution of clusters over time and 2) a hierarchical glyph-based technique that uses data aggregation and small multiples techniques to visualize the propagation of patterns in their spatial domain. ECoG ClusterFlow makes it possible 1) to compare the spatio-temporal evolution patterns for continuous and discontinuous time-frames, 2) to aggregate data to compare and contrast temporal information at varying levels of granularity, 3) to investigate the evolution of spatial patterns without occluding the spatial context information. We present the results of our case studies on both simulated data and real epileptic seizure data aimed at evaluating the effectiveness of our approach in discovering meaningful patterns and insights to the domain experts in our team.
numpy networkx community PySide Consensus Clustering ################################
Note: Tested on OS X 10.11.6 and Ubuntu 14.04
# Conda installation, if you dont have conda, then install via (http://conda.pydata.org/docs/install/quick.html)
#Download ECoG Cluster repository and then goto src folder
#For mac switch to macports python for installation (as installation libraries are in macports):
sudo port install python27
port select --list python
sudo port select --set python python27
#For mac set the pip for macports:
sudo port install py27-pip
sudo port select --list pip
sudo port select --set pip pip27
sudo pip install numpy==1.11.0
sudo pip install networkx==1.11
sudo pip install nibabel==2.0.2
sudo pip install pydotplus
sudo pip install python-louvain
To install QT:
for linux
sudo apt-get install libqt4-dev
sudo easy_install -U PySide
for mac
sudo port install qt4-mac
sudo port install py27-pyside
#Download ECoG Cluster repository and then goto src folder
ecogClusterdir/src> RunProjectMain.py
Happy Analysis!
####Major Files BrainViewerDataPathsArtificial.py -- path for the dataset
**RunProjectMain2.py** -- path for running the application
##Running the Tool ecogClusterdir/src> python RunProjectMain2.py
See Contributing
"ECoG Cluster Flow” Copyright (c) 2017, The Regents of the University of California, through Lawrence Berkeley National Laboratory (subject to receipt of any required approvals from the U.S. Dept. of Energy). All rights reserved.
Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met:
(1) Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer.
(2) Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution.
(3) Neither the name of the University of California, Lawrence Berkeley National Laboratory, U.S. Dept. of Energy nor the names of its contributors may be used to endorse or promote products derived from this software without specific prior written permission.
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT OWNER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
You are under no obligation whatsoever to provide any bug fixes, patches, or upgrades to the features, functionality or performance of the source code ("Enhancements") to anyone; however, if you choose to make your Enhancements available either publicly, or directly to Lawrence Berkeley National Laboratory, without imposing a separate written license agreement for such Enhancements, then you hereby grant the following license: a non-exclusive, royalty-free perpetual license to install, use, modify, prepare derivative works, incorporate into other computer software, distribute, and sublicense such enhancements or derivative works thereof, in binary and source code form.
Please cite ECoG ClusterFlow in your publications if it helps your research:
@Article{Murugesan2017,
author="Murugesan, Sugeerth
and Bouchard, Kristofer
and Chang, Edward
and Dougherty, Max
and Hamann, Bernd
and Weber, Gunther H.",
title="Multi-scale visual analysis of time-varying electrocorticography data via clustering of brain regions",
journal="BMC Bioinformatics",
year="2017",
volume="18",
number="6",
pages="236",
doi="10.1186/s12859-017-1633-9",
}
Our conference work:
@inproceedings{Murugesan:2016:HSV:2975167.2985688,
author = {Murugesan, Sugeerth and Bouchard, Kristofer and Chang, Edward and Dougherty, Max and Hamann, Bernd and Weber, Gunther H.},
title = {Hierarchical Spatio-temporal Visual Analysis of Cluster Evolution in Electrocorticography Data},
booktitle = {Proceedings of the 7th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics},
series = {BCB '16},
year = {2016},
isbn = {978-1-4503-4225-4},
location = {Seattle, WA, USA},
pages = {630--639},
numpages = {10},
url = {http://doi.acm.org/10.1145/2975167.2985688},
doi = {10.1145/2975167.2985688},
acmid = {2985688},
publisher = {ACM},
address = {New York, NY, USA},
keywords = {Brain Imaging, Electrocorticography, Graph Visualization, Linked Views, Neuroinformatics},
}
The ECoG ClusterFlow Project makes use of the following libraries
- Numpy, Copyright (C) 2004-2016, NetworkX is distributed with the BSD license
- PYTHON LOUVAIN, Copyright (c) 2009, Thomas Aynaud thomas.aynaud@lip6.fr
- PYSIDE, Copyright (C) 1991, 1999 Free Software Foundation, Inc , GNU LESSER GENERAL PUBLIC LICENSE
- Scipy, Copyright © 2001, 2002 Enthought, Inc.
- Consensus Clustering, Copyright Michael Seiler 2008, Rutgers University, miseiler@gmail.com