Spatial feature fusion in 3D convolutional architectures for lung tumor segmentation from 3D CT images
Publication
Najeeb, S., & Bhuiyan, M. I. H. (2022). Spatial feature fusion in 3D convolutional autoencoders for lung tumor segmentation from 3D CT images. Biomedical Signal Processing and Control, 78, 103996.
Read Online | PDF | preprint
Abstract
Accurate detection and segmentation of lung tumors from volumetric CT scans is a critical area of research for the development of computer aided diagnosis systems for lung cancer. Several existing methods of 2D biomedical image segmentation based on convolutional autoencoders show decent performance for the task. However, it is imperative to make use of volumetric data for 3D segmentation tasks. Existing 3D segmentation networks are computationally expensive and have several limitations. In this paper, we introduce a novel approach which makes use of the spatial features learned at different levels of a 2D convolutional autoencoder to create a 3D segmentation network capable of more efficiently utilizing spatial and volumetric information. Our studies show that without any major changes to the underlying architecture and minimum computational overhead, our proposed approach can improve lung tumor segmentation performance by 1.61%, 2.25%, and 2.42% respectively for the 3D-UNet, 3D-MultiResUNet, and Recurrent-3D-DenseUNet networks on the LOTUS dataset in terms of mean 2D dice coefficient. Our proposed models also respectively report 7.58%, 2.32%, and 4.28% improvement in terms of 3D dice coefficient. The proposed modified version of the 3D-MultiResUNet network outperforms existing segmentation architectures on the dataset with a mean 2D dice coefficient of 0.8669. A key feature of our proposed method is that it can be applied to different convolutional autoencoder based segmentation networks to improve segmentation performance.
Citation
@article{NAJEEB2022103996,
title = {Spatial feature fusion in 3D convolutional autoencoders for lung tumor segmentation from 3D CT images},
journal = {Biomedical Signal Processing and Control},
volume = {78},
pages = {103996},
year = {2022},
issn = {1746-8094},
doi = {https://doi.org/10.1016/j.bspc.2022.103996},
url = {https://www.sciencedirect.com/science/article/pii/S174680942200444X},
author = {Suhail Najeeb and Mohammed Imamul Hassan Bhuiyan},
keywords = {Segmentation, CT scan, Lung tumor, Convolutional autoencoders, Deep learning},
abstract = {Accurate detection and segmentation of lung tumors from volumetric CT scans is a critical area of research for the development of computer aided diagnosis systems for lung cancer. Several existing methods of 2D biomedical image segmentation based on convolutional autoencoders show decent performance for the task. However, it is imperative to make use of volumetric data for 3D segmentation tasks. Existing 3D segmentation networks are computationally expensive and have several limitations. In this paper, we introduce a novel approach which makes use of the spatial features learned at different levels of a 2D convolutional autoencoder to create a 3D segmentation network capable of more efficiently utilizing spatial and volumetric information. Our studies show that without any major changes to the underlying architecture and minimum computational overhead, our proposed approach can improve lung tumor segmentation performance by 1.61%, 2.25%, and 2.42% respectively for the 3D-UNet, 3D-MultiResUNet, and Recurrent-3D-DenseUNet networks on the LOTUS dataset in terms of mean 2D dice coefficient. Our proposed models also respectively report 7.58%, 2.32%, and 4.28% improvement in terms of 3D dice coefficient. The proposed modified version of the 3D-MultiResUNet network outperforms existing segmentation architectures on the dataset with a mean 2D dice coefficient of 0.8669. A key feature of our proposed method is that it can be applied to different convolutional autoencoder based segmentation networks to improve segmentation performance.}
}
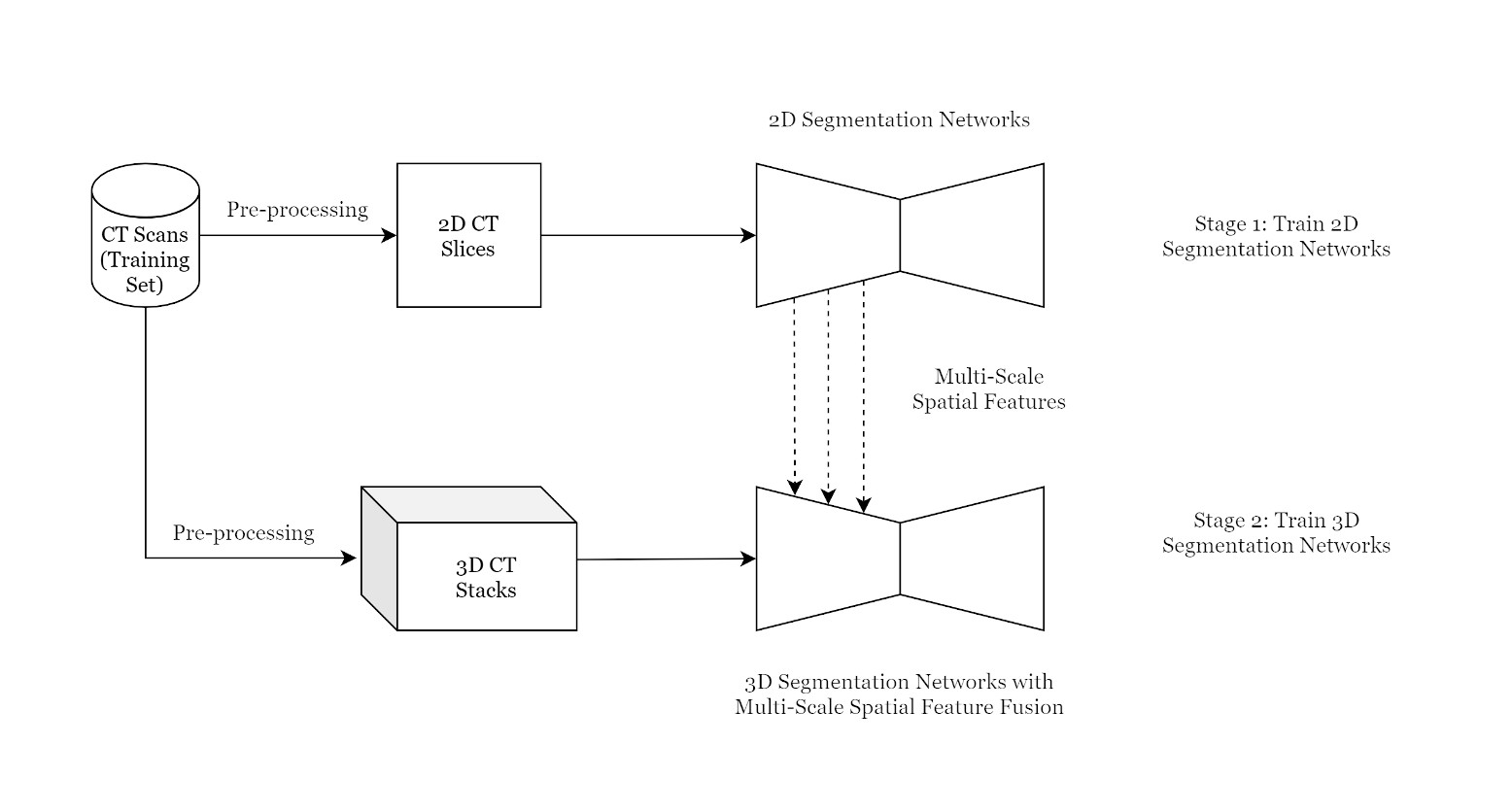
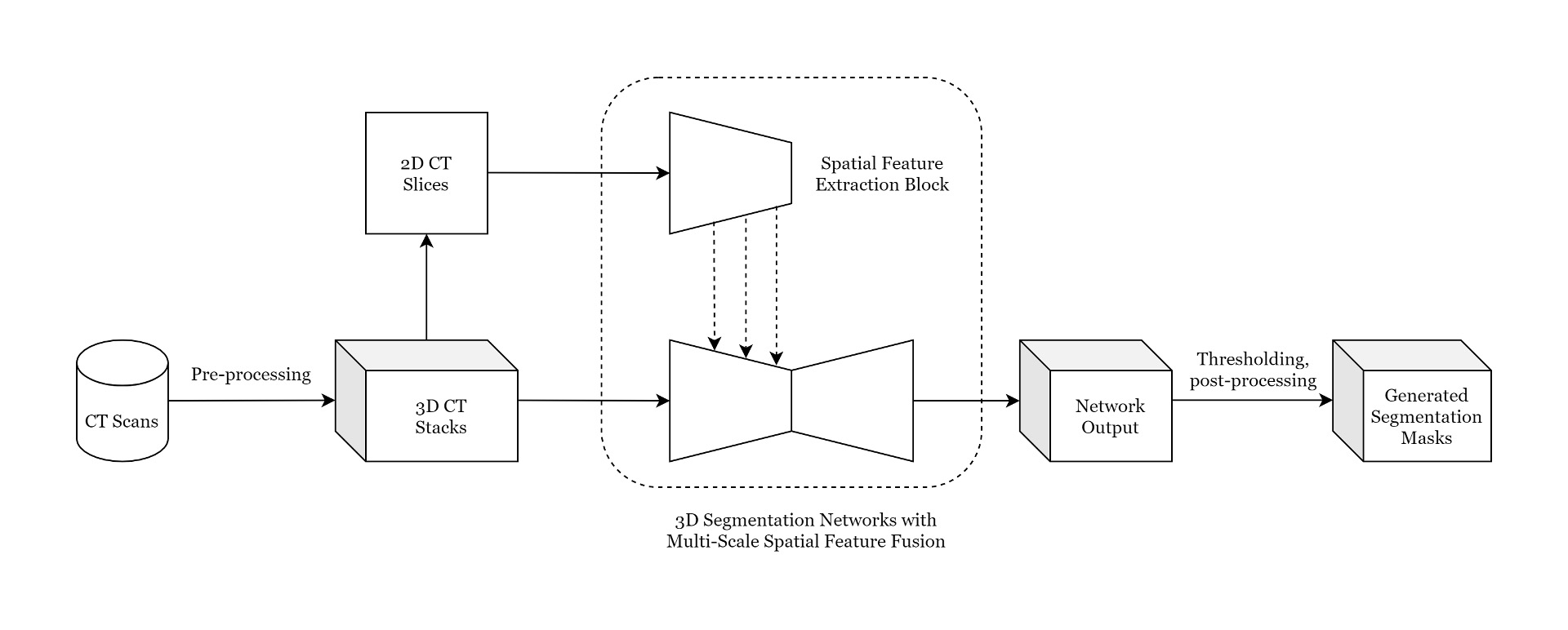
Proposed Methodology
Dataset
LOTUS Benchmark (Lung-Originated Tumor Region Segmentation)
- Prepared as part of the IEEE VIP Cup 2018 Challenge
- Modified version of NSCLC-Radiomics Dataset
- Contains Computed Tomography (CT) scans of 300 lung cancer patients with NSCLC.
- CT Scan Resolution: 512x512
Segmentation Task:
- GTV (Gross-Tumor Volume)
Dataset Statistics:
| CT Scanner | Number of Slices | ||||
|---|---|---|---|---|---|
| Dataset | Patients | CMS | Siemens | Tumor | Non-Tumor |
| Train | 260 | 60 | 200 | 4296(13.7%) | 26951(86.3%) |
| Test | 40 | 34 | 6 | 848(18.9%) | 3610(81.1%) |
Data Processing
- Read scans: PyDicom Library
- Normalize HU Values between 0~1
- Slices resized to resolution 256x256
Sample Scans:
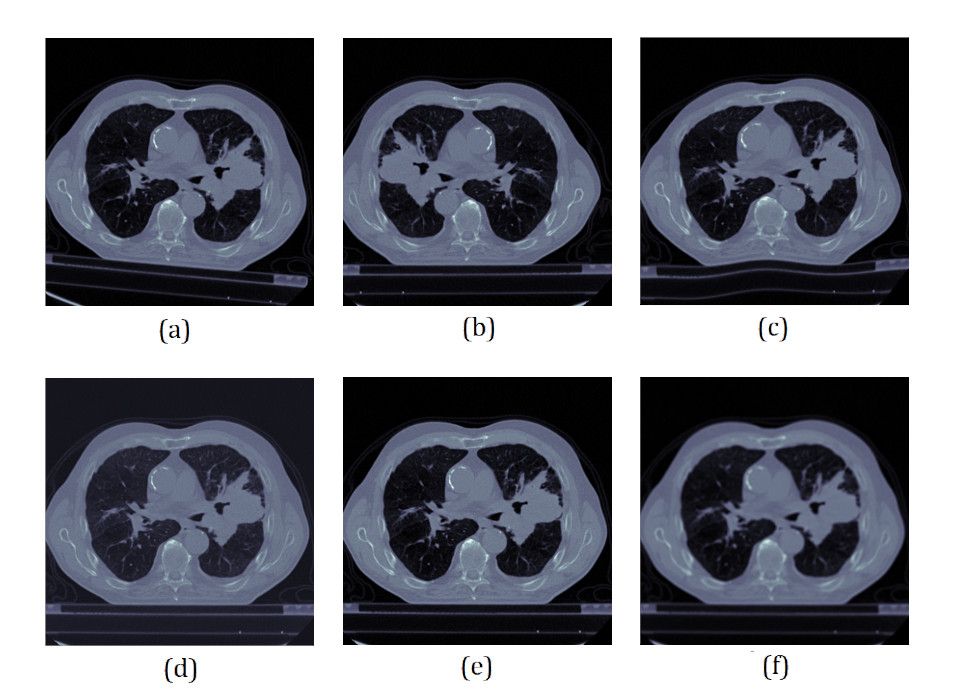
Data Augmentation
Data augmemntation is performed on-the-fly. One or more of the following data augmantations are performed on each training samples -
(a) Random Rotation (b) Horizontal Flip (c) Random Elastic Deformation (d) Random Contrast Normalization (e) Random Noise (f) Blurring
Illustration:
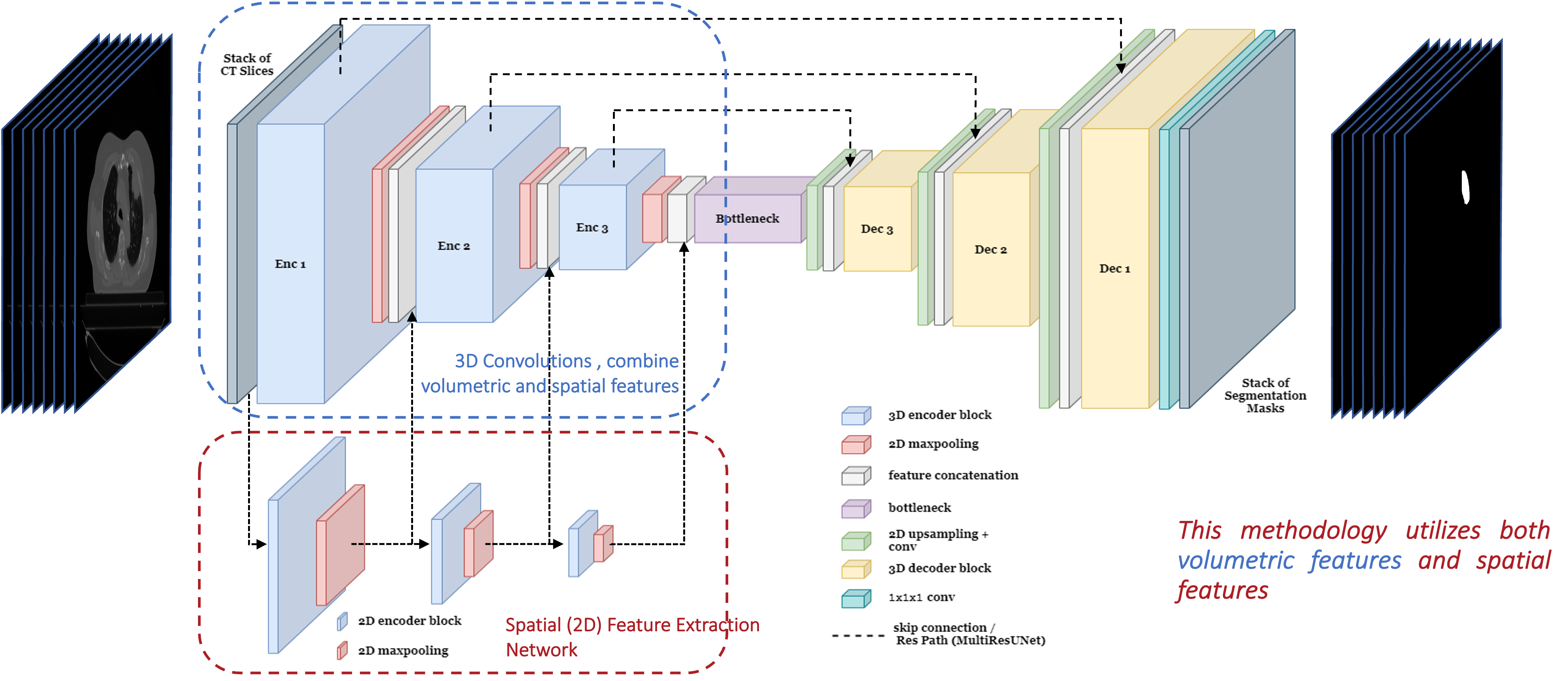
Baseline Architectures
- UNet (3D)
- MultiResUNet (3D)
- Recurrent-3D-DenseUNet
Multi-Scale Spatial Feature Extractor:
The encoders of the 2D segmentation networks are utilized to extract spatial (2D) features from 2D CT slices.
Proposed Pipeline
Architecutre:
Mask Generation
- 3D Segmentation networks: produce results on 8 consecutive slices
- To produce final segmentation masks, overlapping stacks of CT slices are processed by the segmentation networks.
- Overlapping masks are averaged which serves as a post-processing step to remove noise.
- Segmentation mask values are within 0~1 where 0 signifies no tumor and 1 signifies tumor
- Two-step thresholding approach is applied to generate final segmentation mask
- Step 1: Apply a threshold of 0.7 to filter out false-positive slices
- Step 2: Apply a threshold of 0.5 to generate the final tumor volume
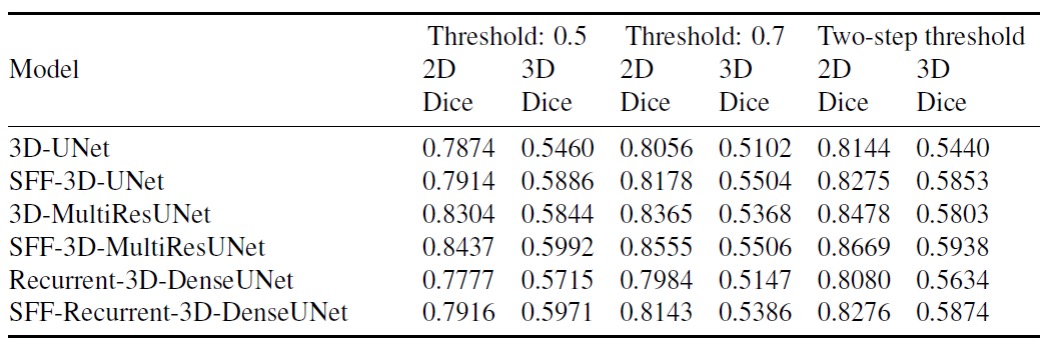
Results
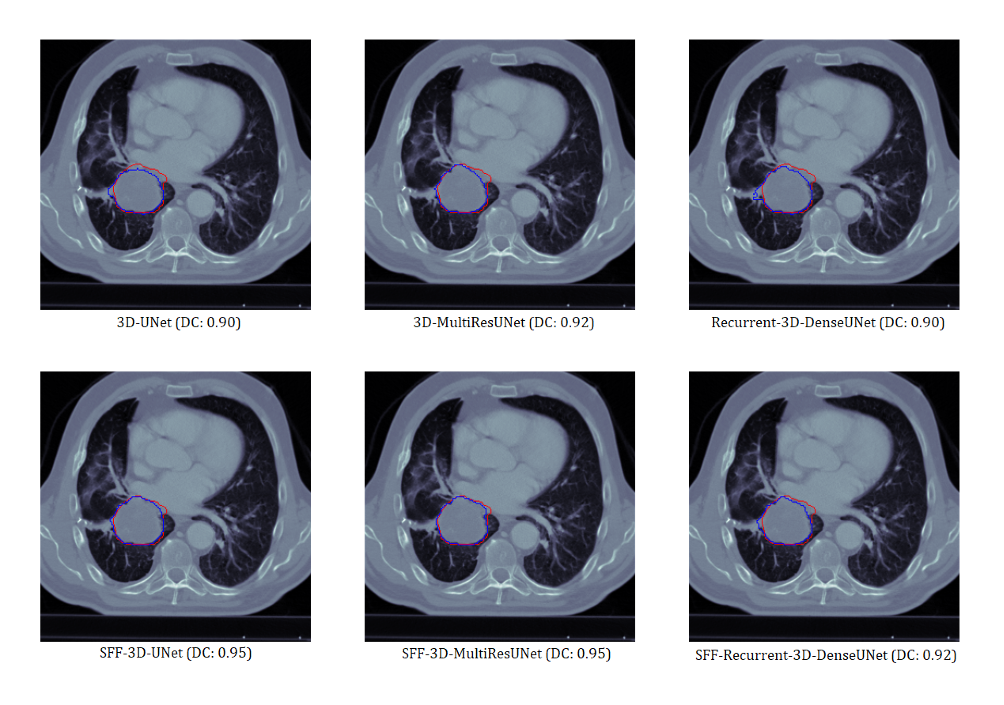
In terms of 2D dice coefficient, the proposed models with SFF achieve performance improvements of -
| 2D Dice Score | Without SFF | With SFF | Improvement |
|---|---|---|---|
| 3D-UNet | 0.8144 | 0.8275 | 1.61% |
| 3D-MultiResUNet | 0.8478 | 0.8669 | 2.25% |
| Recurrent-3D-DenseUNet | 0.8080 | 0.8276 | 2.42% |
In terms of 3D dice coefficient, the proposed models with SFF achieve performance improvements of -
| 3D Dice Score | Without SFF | With SFF | Improvement |
|---|---|---|---|
| 3D-UNet | 0.5440 | 0.5853 | 7.58% |
| 3D-MultiResUNet | 0.5803 | 0.5938 | 2.32% |
| Recurrent-3D-DenseUNet | 0.5634 | 0.5874 | 4.28% |
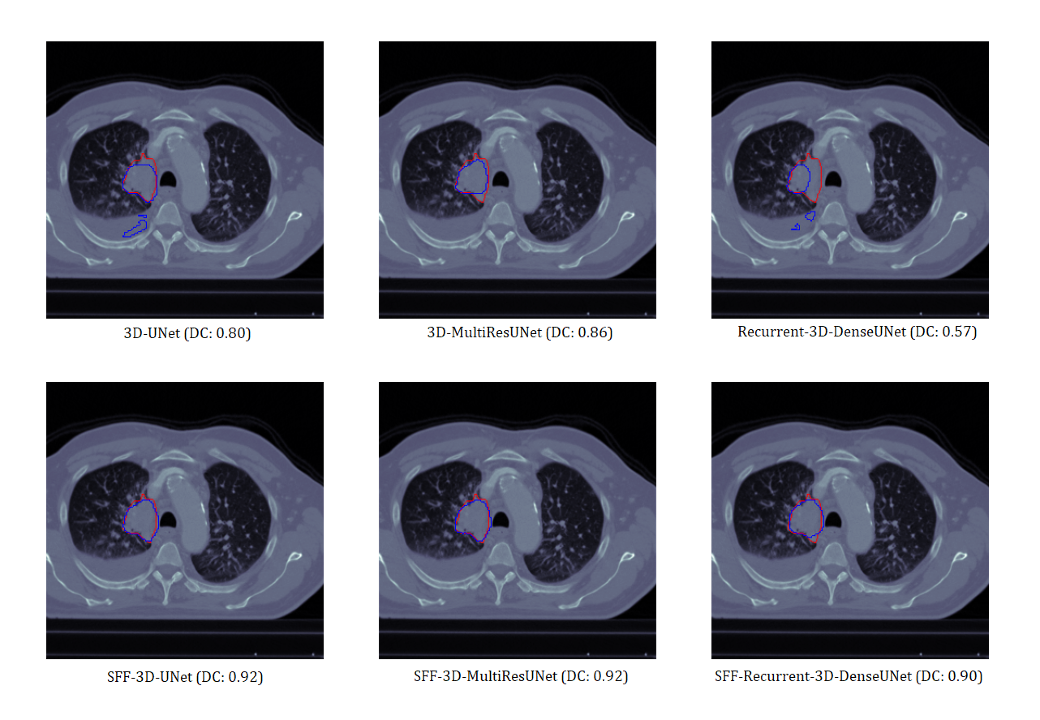
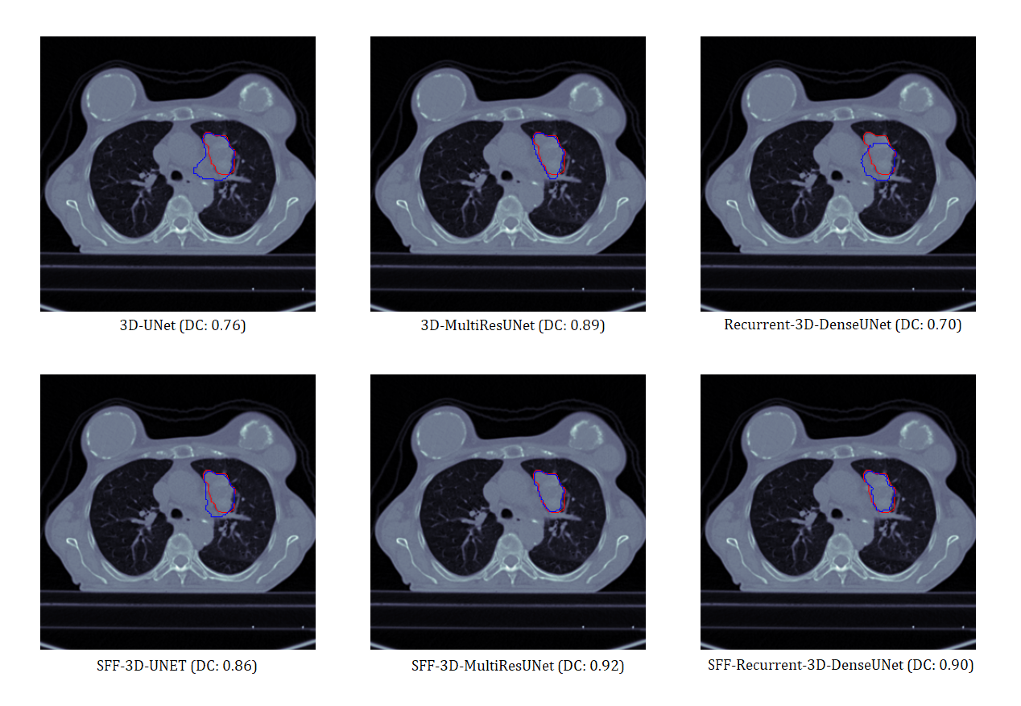
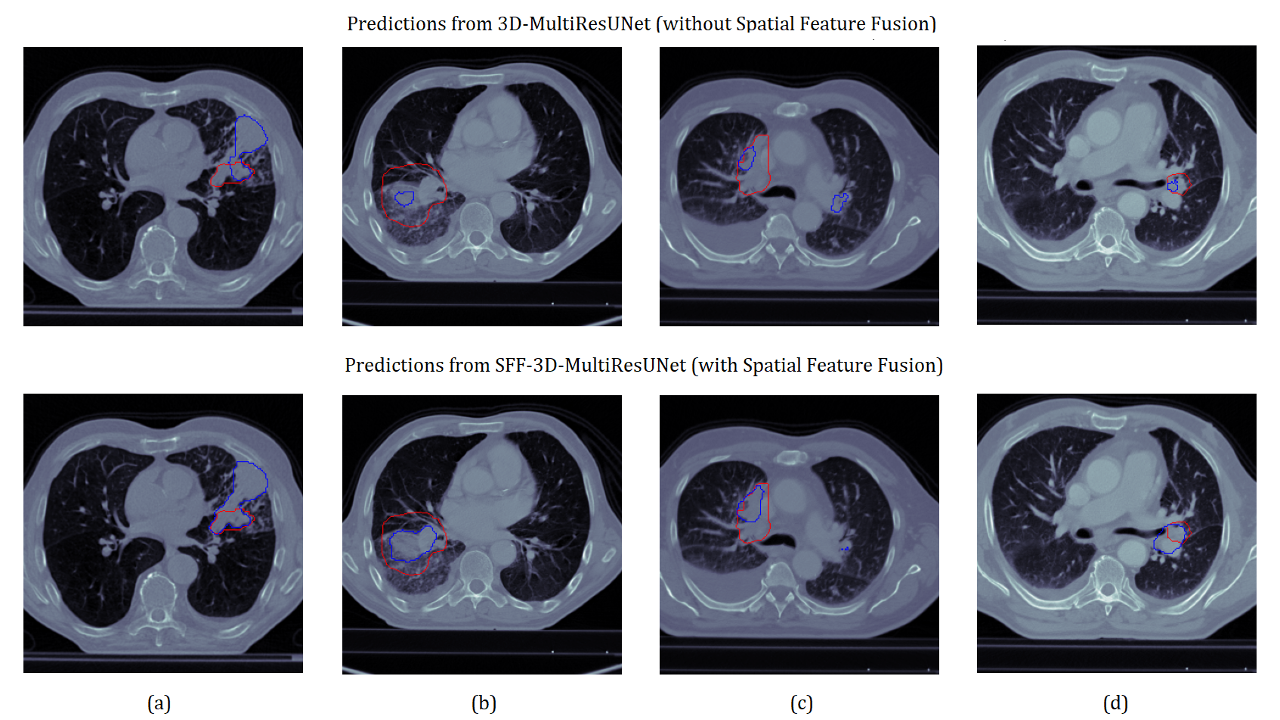
Visual Analysis
Sample 1
Sample 2
Edge Cases
Summary
- We have proposed three novel architecutres which incorporate multi-scale spatial feature fusion and improve lung tumor segmentation performance with minimal computational overhead.
- Our proposed architectures achieved performance improvements of 1.61%, 2.25%, and 2.42% respectively in terms of 2D dice coefficient.
- Our proposed architectures also achieved performance improvements of 7.58%, 2.32%, and 4.28% in terms of 3D dice coefficient.
Source Code
To obtain the baseline 2D models train using the script code/train_2d.py. The 3D data generation/pipeline can be found in here - https://github.com/muntakimrafi/TIA2020-Recurrent-3D-DenseUNet , https://github.com/udaykamal20/Team_Spectrum which is the implementation of the Recurrent-3D-DenseUNet. The models used in this publication can be found in code/model_lib.py. Example usage of the models with multi-scale-spatial-feature-fusion:
def get_HybridUNet001(model_fe, input_shape = (256, 256, 8, 1)):
'''
Usage:
base_model = load_model('preferred type: UNet2Dx3 (functional)')
model_fe = get_model_fe(base_model)
model = get_HybridUNet001(model_fe)
'''
pass