- This repository contains the end-to-end analysis workflow of phenotypic data. The workflow is divided into two parts: 1) Pre-processing and Quality Check and 2) Data Analysis analysis in ASReml R package and lme4 R package.
- In this pipeline we demosntrated the state-of-the-art implementation of the phenotypic data analysis pipeline and workflow embedded into a well-descriptive document.
- The developed analytical pipeline is open-source, demonstrating how to analyze the phenotypic data in crop breeding programs with step-by-step instructions.
- The analysis pipeline shows how to pre-process and check the quality of phenotypic data, perform robust data analysis using modern statistical tools and approaches, and convert it into a reproducible document.
- Explanatory text with R codes, outputs either in text, tables, or graphics, and interpretation of results are integrated into the unified document. The analysis is highly reproducible and can be regenerated at any time.
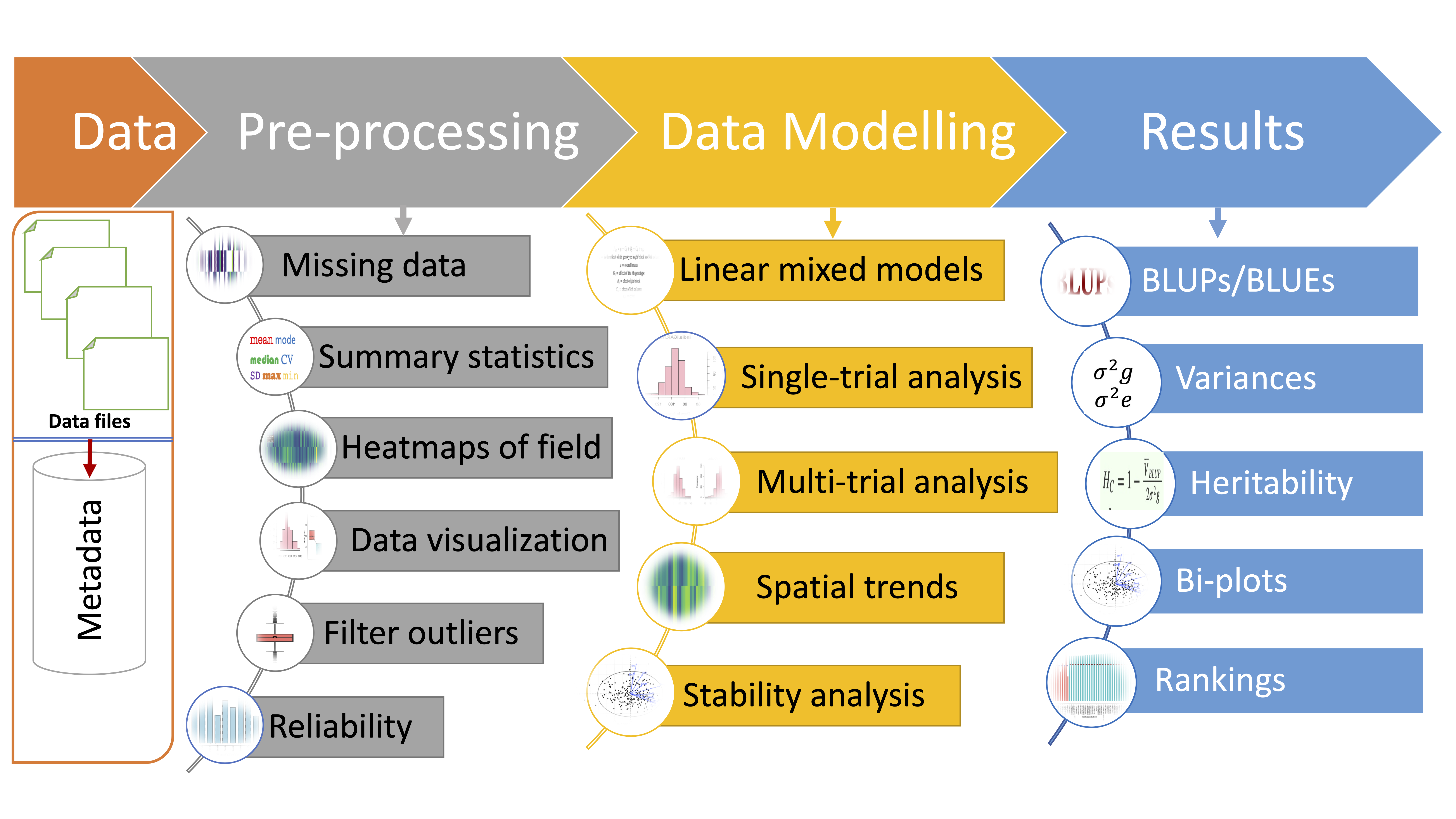
- The schematic representation of analysis workflow is given in figure:
-
The source in .Rmd is available here Sample Pre Processing
-
The HTML file is available here Pre-processing and Quality Check HMTL
- Data analysis is shown both in ASReml R package and in lme4 R package. The reason to show analysis in lme4 R package is because it is free and open source R package and can be used by all users. For ASReml R users need license.
-
The source in .Rmd is available here Asreml R Workflow
-
The HTML file is available here ASReml R Workflow HMTL
- The source in .Rmd is available here lme4 R Workflow
- The HTML file is available here lme4 R Workflow HTML
The steps to use the source codes and run it on local computer is given below:
-
Open the GitHub page containing the source codes and files by using clicking the link https://github.com/whussain2/Analysis-pipeline.git.
-
This will pop-up the Github repository, click on the Code button on right side of page highlighted as green box and scroll and click on Download Zip.
-
Save and unzip the downloaded repository in local drive.
-
Open the .Rmd file in R Studio and make sure to change the working directory based on users defined path to the repository.
-
Also users must install and upload the following R packages before running the pipeline:
library(easypackages)
libraries("dplyr", "reshape2", "readxl", "ggpubr","stringr", "ggplot2",
"tidyverse","lme4", "data.table", "readr","plotly", "DT",
"pheatmap","asreml", "VennDiagram", "patchwork", "heatmaply",
"ggcorrplot", "RColorBrewer", "hrbrthemes", "tm", "proustr", "arm",
"gghighlight", "desplot", "gridExtra", "TeachingDemos", "scales", "ASExtras4",
"FactoMineR", "corrplot", "factoextra", "asremlPLUS")
Note: The pipeline requires asreml R package to do the analysis. If you dont have license for ASReml R then use lme4 R workflow to perform analysis.
More details can be found in the manuscript available at Link
Rainfed Breeding Team IRRI
You may contact the author of this code, Waseem Hussain at waseem.hussain@irri.org; waseemhussain907@gmail.com