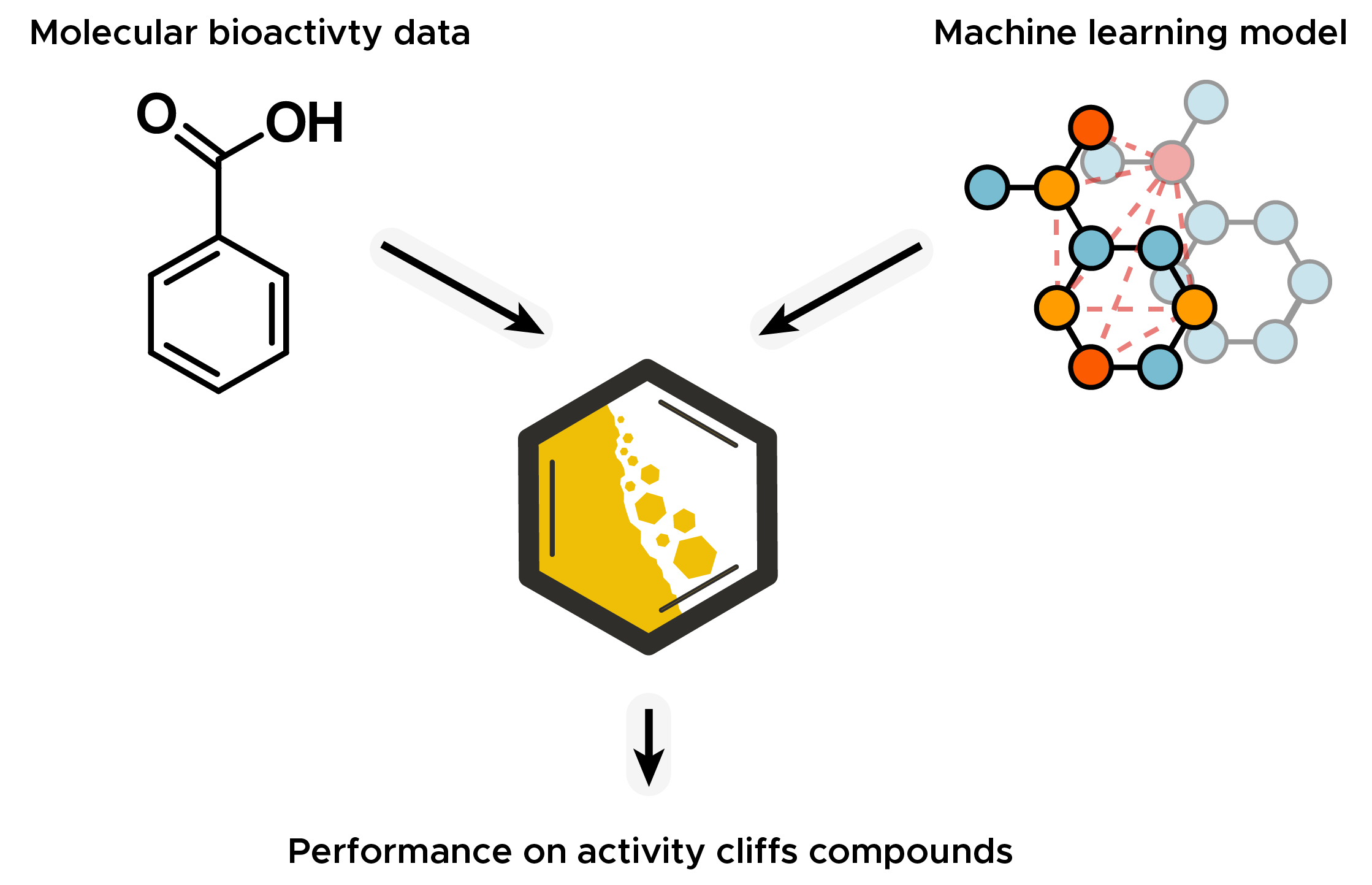
Molecule Activity Cliff Estimation (MoleculeACE) is a tool for evaluating the predictive performance on activity cliff compounds of machine learning models.
MoleculeACE can be used to:
- Analyze and compare the performance on activity cliffs of machine learning methods typically employed in QSAR.
- Identify best practices to enhance a model’s predictivity in the presence of activity cliffs.
- Design guidelines to consider when developing novel QSAR approaches.
Table of Contents
In a benchmark study we collected and curated bioactivity data on 30 macromolecular targets, which were used to evaluate the performance of many machine learning algorithms on activity cliffs. We used classical machine learning methods combined with common molecular descriptors and neural networks based on unstructured molecular data like molecular graphs or SMILES strings.
Activity cliffs are molecules with small differences in structure but large differences in potency. Activity cliffs play an important role in drug discovery, but the bioactivity of activity cliff compounds are notoriously difficult to predict.
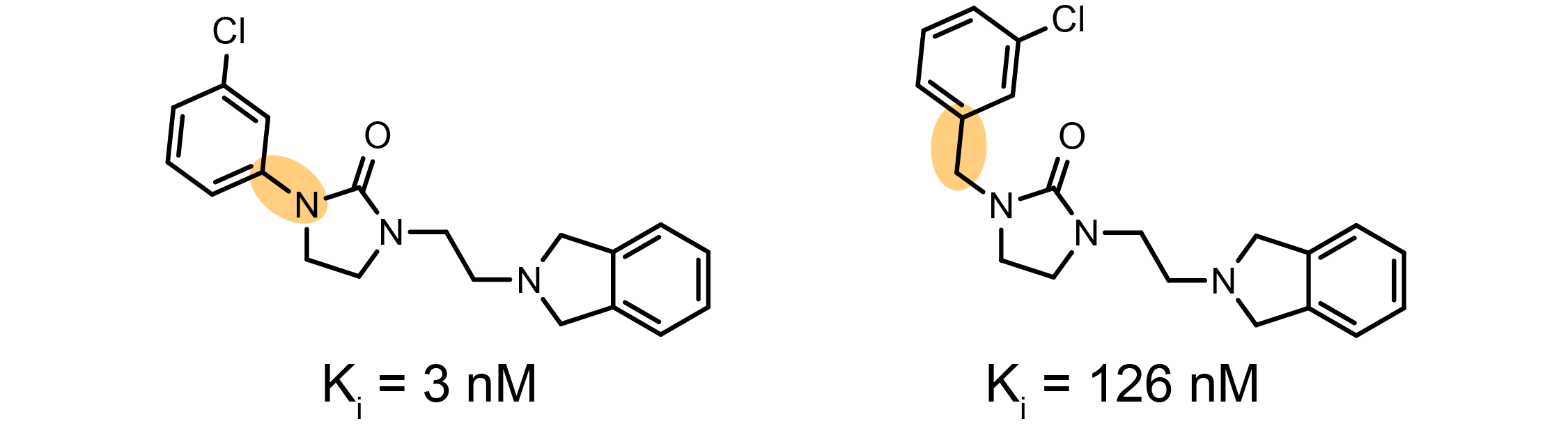 Example of an activity cliff on the Dopamine D3 receptor, D3R
Example of an activity cliff on the Dopamine D3 receptor, D3R
Any regression model can be evaluated on activity cliff performance using MoleculeACE on third party data or the 30 included molecular bioactivity data sets. All 24 machine learning strategies covered in our benchmark study can be used out of the box.
MoleculeACE currently supports Python 3.8. Some required deep learning packages are not included in the pip install.
- Tensorflow (2.9.0)
- PyTorch (1.11.0)
- PyTorch Geometric (2.0.4)
- Transformers (4.20.1)
pip install MoleculeACE
git clone https://github.com/derekvantilborg/MoleculeACE
pip install rdkit-pypi pandas numpy pandas chembl_webresource_client scikit-learn matplotlib tqdm python-Levenshtein
from MoleculeACE import MPNN, Data, Descriptors, calc_rmse, calc_cliff_rmse, get_benchmark_config
dataset = 'CHEMBL2034_Ki'
descriptor = Descriptors.GRAPH
algorithm = MPNN
# Load data
data = Data(dataset)
# Get the already optimized hyperparameters
hyperparameters = get_benchmark_config(dataset, algorithm, descriptor)
# Featurize SMILES strings with a specific method
data(descriptor)
# Train and a model
model = algorithm(**hyperparameters)
model.train(data.x_train, data.y_train)
y_hat = model.predict(data.x_test)
# Evaluate your model on activity cliff compounds
rmse = calc_rmse(data.y_test, y_hat)
rmse_cliff = calc_cliff_rmse(y_test_pred=y_hat, y_test=data.y_test, cliff_mols_test=data.cliff_mols_test)
print(f"rmse: {rmse}")
print(f"rmse_cliff: {rmse_cliff}")from MoleculeACE import calc_rmse, calc_cliff_rmse
# Train your own model
model = ...
y_hat = model.predict(...)
# Evaluate your model on activity cliff compounds
rmse = calc_rmse(y_test, y_hat)
# You need to provide both the predicted and true values of the test set + train labels + the train and test molecules
# Activity cliffs are calculated on the fly
rmse_cliff = calc_cliff_rmse(y_test_pred=y_hat, y_test=y_test, smiles_test=smiles_test, y_train=y_train,
smiles_train=smiles_train, in_log10=True, similarity=0.9, potency_fold=10)
print(f"rmse: {rmse}")
print(f"rmse_cliff: {rmse_cliff}")You can currently cite our pre-print:
van Tilborg et al. (2022). Exposing the limitations of molecular machine learning with activity cliffs. ChemRxiv.
MoleculeACE is under MIT license. For use of specific models, please refer to the model licenses found in the original packages.