Adversarially Robust Prototypical Few-shot Segmentation with Neural-ODEs
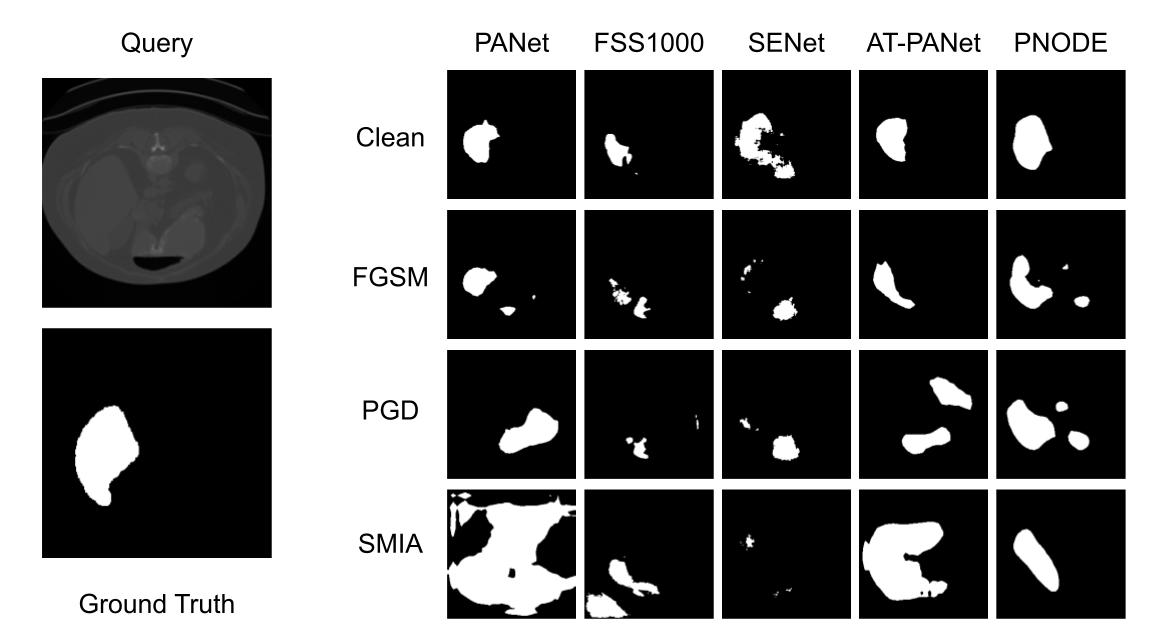
Predicted masks by different models for different attacks
To install this repository and its dependent packages, run the following.
git clone https://github.com/pnodemiccai/PNODE-FSS.git
cd PNODE-FSS
conda create --name PNODE-FSS # (optional, for making a conda environment)
pip install -r requirements.txt
Change the paths to BCV, CT-ORG and Decathlon datasets in config.py and test_config.py according to paths on your local. Also change the path to ImageNet pretrained VGG model weights in these files.
Trained model weights for PNODE, AT-PANet and all baselines on relevant settings can be found here.
Processed datasets of BCV, CT-ORG and Decathlon can be downloaded from here.
To train baseline methods, go to their respective folders and run
python3 train.py with model_name=<save-name> target=<test-target> n_shot=<shot>
For AT-PANet, run train_adversarial.py from PANet folder with the same arguments as before.
For PNODE, some extra arguments are needed. To train PNODE, run
python3 train.py with model_name=<save-name> target=<test-target> n_shot=<shot> ode_layers=3 ode_time=4
To train baseline methods, go to their respective folders and run
python3 test_attacked.py with snapshot=<weights-path> target=<test-target> dataset=<BCV/CTORG/Decathlon> attack=<Clean/FGSM/PGD/SMIA> attack_eps=<eps> to_attack=<q/s>
This command can be used for testing all models on all settings, namely 1-shot and 3-shot, liver and spleen and Clean, FGSM, PGD and SMIA with different epsilons.
BCV:
Liver: 6
Spleen: 1
CT-ORG:
Liver: 1
Decathlon:
Liver: 2
Spleen: 6