This repo contains code for the paper "Deep learning enables image-based tree counting, crown segmentation and height prediction at national scale", which is published by PNAS Nexus.
Check the paper at: https://doi.org/10.1093/pnasnexus/pgad076.
run main1_multitask_counting_segmentation.py (bash script: run_script.sh)
--- set configs ---
case1: same input spatial resolution: use config/UNetTraining.py
case2: inputs of different spatial resolution (only support 1/2 coarser resolution for now): use config/UNetTraining_multires.py
--- example data for demonstration ---
example input data in: example_extracted_data/
IMPORTANT: LISCENCE REQUIRED FOR FORMAL USE OF THE DATA!
--- major tunable hyperparameters ---
boundary weights: determine the penalty on crown boundaries for better individual separation
task_ratio: the ratio for weighting the two losses to balance the emphasis on both tasks during training, may differ from case to case
normalize: ratio to augment data by randomly normalize image patches
run main2_height_prediction.py
set configs in config/UNetTraining_CHM.py
run step3_predict_segmentation_counting.py
--- set configs ---
config/Model_compare_multires.py
--- Example prediction ---
See /example_extracted_data/
segmentation result: seg_41.tif
counting result: density_41.tif
--- Note that the model was trained using image patch no.41, and thus should not be tested using the same image in the test phrase. Here we simply demonstrate how to apply the model on a test image. ---
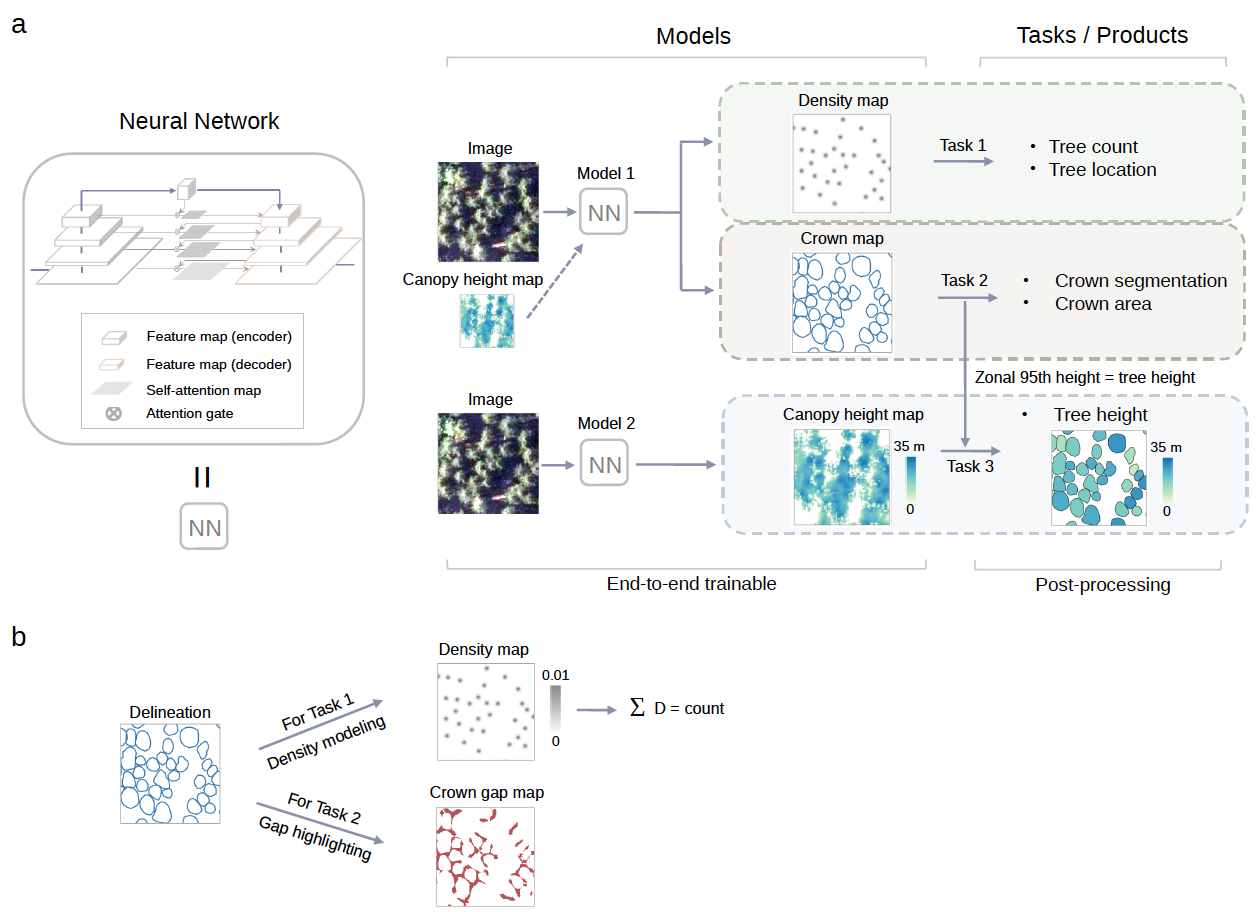
Figure 1. Overview of the framework used to count individual trees and predict their crown area and height. a, Deep learning-based framework for individual tree counting, crown segmentation, and height prediction. Spatial locations of individual trees are incorporated in the tree density maps and the crown segmentation maps. The canopy height map (CHM) derived from LiDAR data provides pixel-wise height information, which, when available for a specific study area, can optionally be used as an additional input band for the individual tree counting and crown segmentation tasks. b, Data preparation and modeling for tree counting and crown segmentation. The manually delineated individual tree crowns are modeled as density maps for the counting task by extracting the polygon centroids. The gaps between adjacent crowns are highlighted for the separation of individual tree crowns during the training phase.
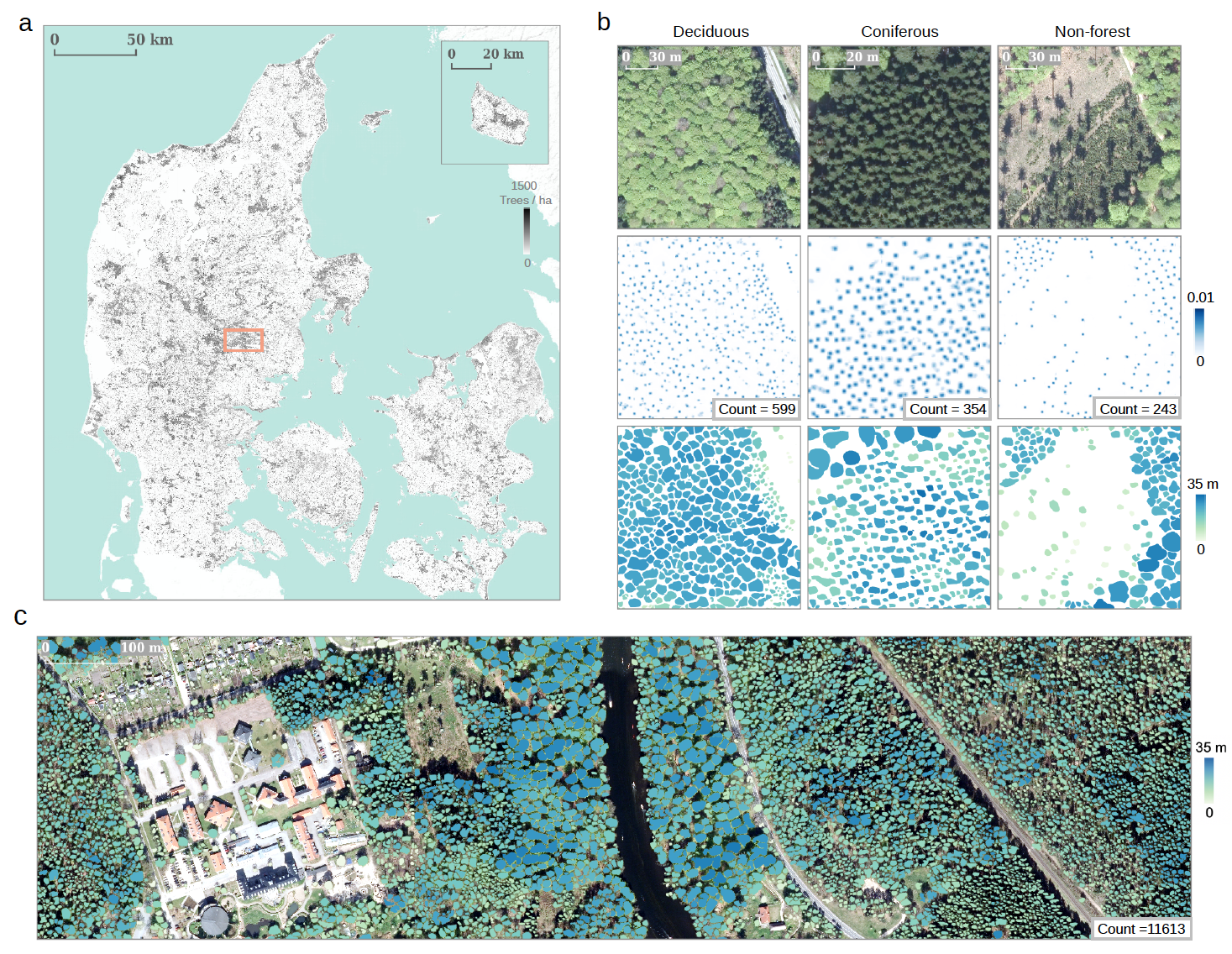
Figure 2. Example products from the proposed framework. a, Wall-to-wall tree count prediction for Denmark. b, Detailed examples showing the individual tree counting (second row), crown segmentation (third row), and height prediction (third row) from three major types of landscapes (deciduous forest, coniferous forest, and non-forest). c, Large-scale individual tree crown segmentation results colored by height predictions. Examples in b and c were sampled from the region indicated by the orange box in a.