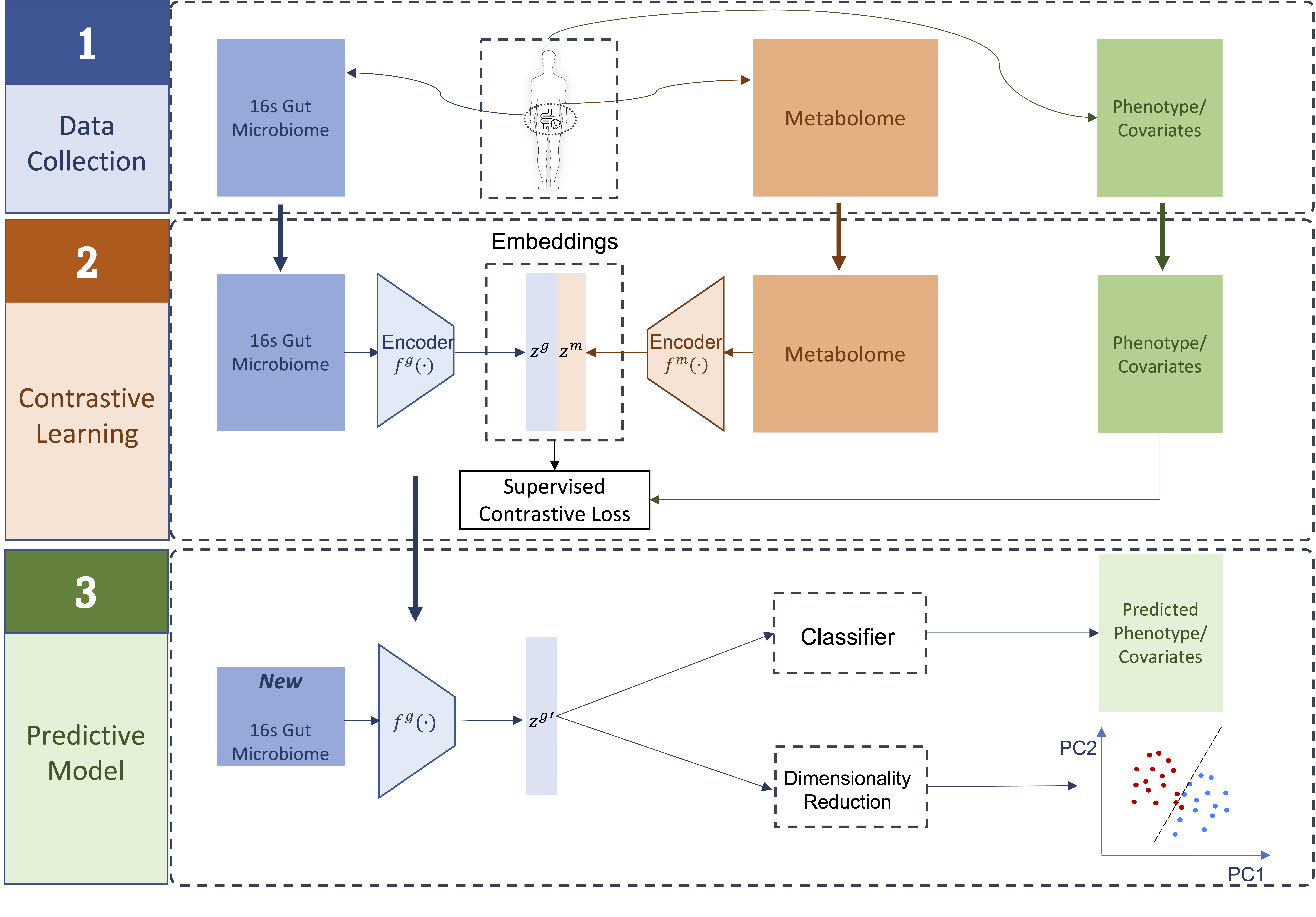
The human microbiome consists of trillions of microorganisms cohabitating a variety of body niches. Microbiota can modulate the host physiology through microbiota-derived molecule and metabolite interactions. Microbiome-based biomarkers have great potential for numerous disease states but current approaches have yielded limited success.
Here, we propose a novel integrative modeling framework, Microbiome-based Supervised Contrastive Learning Framework (MB-SupCon) to improve microbiome-based prediction models. By integrating microbiome and metabolomics data under a supervised contrastive learning scheme, MB-SupCon trained two encoder networks to maximize the similarity between microbiome embedding and metabolomics embedding. Compared to the original microbiome data, the microbiome embedding can lead to improved prediction performances.
Fig.1 MB-SupCon: Microbiome-based Supervised Contrastive Learning Framework
Folders
"data" folder includes:
-
Raw gut 16s microbiome data and metabolome data;
-
Clinical covariates data of all subjects;
-
Index labels for training, validation and testing datasets used for
mixOmicsmethods; -
Output of
mixOmicsmethods (sPLSDA, sPLS and DIABLO).
"tuning" folder includes the Jupyter Notebooks for tuning and result visualization. All tuning results are saved in "tuning/save_tuning" folder;
(Note: detailed tuning results are not included in this repository because of the huge amount and size of the files. You can easily reproduce the tuning results by running notebooks 1b-i - tune with random seeds 1-3.ipynb, 1b-ii - tune with random seeds 4-6.ipynb, 1b-iii - tune with random seeds 7-9.ipynb and 1b-iv - tune with random seeds 10-12.ipynb.)
"embeddings" folder includes feature embedding by MB-SupCon in representation space for all covariates.
"figures" folder includes all loss curves of MB-SupCon and generated lower-dimensional scatter plots by different methods including MB-SupCon.
"models" folder includes trained models.
"outputs" folder includes the output PC1 and PC2 scores (by PCA) used for lower-dimensional scatter plots.
"other_methods" folder includes the codes, plots and outputs of the other methods including MLP, MB-simCLR, mixOmics methods (sPLSDA, sPLS and DIABLO) for comparison with MB-SupCon.
Codes
1a - MB-SupCon_categorical covariates.ipynb: the main Jupyter Notebook used to
- Train MB-SupCon models for different covaraites;
(Note: the best combination of hyperparameters are chosen based on the tuning result. Details can be found in the Jupyter Notebook located at ./tuning/1b-v - save and visualize the tuning results_random seeds_1-12.ipynb.)
-
Output and save corresponding feature embedding in the representation domain;
-
Make predictions of different covariates based on embedding of MB-SupCon and original data, and calculate the average prediction accuracy on testing datasets from multiple training-validation-testing splits;
-
Generate scatter plots on lower-dimensional space by PCA.
(Note: you can choose other dimensionality reduction techniques, such as t-SNE and UMAP).
supervised_loss.py: a function used for calculating supervised contrastive loss;
plotting_utils.py, pred_utils.py, utils_eval.py: utility functions;
mbsupcon.py: the python script for building a MB-SupCon model;
mbsimclr.py: the python script for building a MB-simCLR model.
- System information:
System: Linux; Release: 3.10.0-957.el7.x86_64;
GPU: Tesla V100-SXM2-32GB.
- Version of Python and some important packages used in this repository:
Python version: 3.8.5.
pytorch: 1.7.1 (Build: py3.8_cuda11.0.221_cudnn8.0.5_0);
numpy: 1.19.2;
pandas: 1.2.1;
scikit-learn: 0.23.2;
matplotlib: 3.3.2;
seaborn: 0.11.2.
@article{yang2022mb,
title = {MB-SupCon: Microbiome-based Predictive Models via Supervised Contrastive Learning},
author = {Yang, Sen and Wang, Shidan and Wang, Yiqing and Rong, Ruichen and Kim, Jiwoong and Li, Bo and Koh, Andrew Y and Xiao, Guanghua and Li, Qiwei and Liu, Dajiang J and others},
journal = {Journal of Molecular Biology},
volume = {434},
number = {15},
pages = {167693},
year = {2022},
publisher = {Elsevier}
}
Sen Yang:
senyang@smu.edu | sen.yang@utsouthwestern.edu
Department of Statistical Science
Southern Methodist University
Dallas, TX 75275
Xiaowei Zhan:
xiaowei.zhan@utsouthwestern.edu
Quantitative Biomedical Research Center, Department of Population and Data Sciences
Center for Genetics of Host Defense
University of Texas Southwestern Medical Center
Dallas, TX 75390