Pre-classification of Long-reads for Memory Efficient Taxonomic assignment
Sylvain Riondet, K. Križanović, J. Marić and M, Šikić, Niranjan Nagarajan
NUS/SoC, Biopolis/GIS, Singapore
Tool in active development, any feedback or bug report is welcome, either through github or on twitter
- Segmentation of NCBI RefSeq into clusters
- Building of taxonomic classifiers' indexes for each cluster
Taxonomic classification of mock communities / metagenomic fastq files
- Assignment of long DNA reads (Nanopore/PacBio) to each cluster
- Classification by the classifier with a subset of RefSeq
- Merging of reports
Kraken2 (Derrick E. Wood et al. 2019) and Centrifuge (D.Kim et al. 2016) are currently automated, and any classifier able to build its index on a set of .fna files with a provided taxid should work.
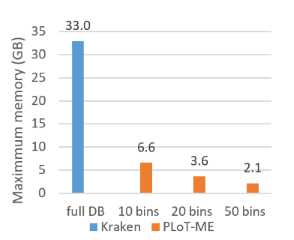
- High reduction in memory needs, defined by the number of clusters *
- Compatible and enhancing existing taxonomic classifiers
- Slight over-head of the pre-classification (currently ~3-5x in time, improvements for future releases)
* Mini Batch K-Means, Web-Scale K-Means Clustering D. Sculley 2010
- Database of Genomes, in .fna / .fasta format, with an associated taxonomy id. Tested with NCBI RefSeq (ftp server)
- Taxonomic classifier, must be installed and added to PATH. Currently supported:
- Kraken2
- Centrifuge
- (feel free to request support for more)
- Linux (tested on Ubuntu 18.04)
- Python >= 3.7
| Package | Version |
|---|---|
| biopython | >= 1.72 |
| ete3 | >= 3.1.1 |
| numpy | >= 1.17.3 |
| pandas | >= 0.23 |
| scikit-learn | >= 0.18 |
| tqdm | >= 4.24.0 |
Create a Python 3 environment with conda
or pyenv.
Installation is then done with pip:
python3 -m pip install plot-me
This will create 2 commands, plot-me.preprocess and plot-me.classify detailed in the 'Usage'.
It is also possible to clone PLoT-ME's repo,
and launching commands directly with python path/to/PLoT-ME/parse_DB.py or classify.py
For the full help: plot-me.preprocess -h
Typical usage:
plot-me.preprocess <path/NCBI/refseq> <folder/for/clusters> <path/taxonomy> -k 4 -w 10000 -n 10 -o <OmitFoldersContainingString>
For the full help: plot-me.classify -h
Typical usage:
plot-me.classify <folder/with/clusters> <folder/reports> -i <fastq files to preclassify>
/mnt/data
|-- mock_files
| |-- mock_community_1.fastq
| | \-- minikm_b10_k3_s10000_oplant-vertebrate (one tmp file per cluster, generated by PLoT-ME)
| \-- mock_community_2.fastq
|-- PLoT-ME
| |-- k3_s10000
| | | -- kmer_counts
| | | |-- counts.k3_s10000 (same tree as RefSeq, with <sequencing_name>.3mer_count.pd)
| | | \-- all-counts.k3_s10000_oplant-vertebrate.csv
| | | -- minikm_b10_k3_s10000_oplant-vertebrate <*>
| | | |-- centrifuge (10 folders with indexes)
| | | |-- kraken2 (10 folders with indexes)
| | | |-- RefSeq_binned (10 folders with fna files)
| | | |-- model.minikm_b10_k3_s10000_oplant-vertebrate.pkl
| | | \-- segments-clustered.minikm_b10_k3_s10000_oplant-vertebrate.pd
| | \ -- minikm_b20_k3_s10000_oplant-vertebrate
| | \-- (same structure)
| |-- k4_s10000
| | ` -- (same structure)
| \-- no-binning
| |-- oAllRefSeq
| \-- oplant-vertebrate
| |-- centrifuge
| \-- kraken2
|-- NCBI
| \-- refseq
|-- reports
| \-- mock_community_1 (one report per cluster)
\-- taxonomy
This <*> can be generated with:
plot-me.preprocess /mnt/data/NCBI/refseq /mnt/data/PLoT-ME /mnt/data/taxonomy -k 3 -w 10000 -n 10 -o plant vertebrate
And can be used with:
plot-me.classify /mnt/data/PLoT-ME/k3_s10000/minikm_b10_k3_s10000_oplant-vertebrate /mnt/data/reports -i /mnt/data/mock_files/mock_community_1.fastq
Python 3 is the main programming language, with extensive use of libraries. Dependencies are resolved using PIP
Data is saved as pickle .pkl or Pandas DataFrame .pd
- Kmer counts Pandas DataFrames are saved under
.../kmer_counts/counts.<param>and have the following columns:
taxon category start end name description fna_path AAAA ... TTTT - Cluster assignments
segments-clustered.\<param\>.pdtrade the nucleotides columns to aclustercolumn. RefSeq_binnedis the clustering made by PLoT-ME, and holds one folder per cluster, with concatenated segments of genomes (one .fna file per taxa)- Libraries generated by classifier, depends on each of them.
The model*.pkl and the folder kraken2 or centrifuge are needed for PLoT-ME to work. Folder tree needs to remain intact.
April 2021
- Implementation of Cython version of the kmer counter
- Adding reverse complement to forward strand
July 2020:
pre-processUsing large k (5+) and small s (10000-) yield very large kmer counts, costing high amounts of RAM (esp. when combining all kmer counts together, RAM needs to reach ~30GB or more).classifyMerging of reportspre-processCleaning of pre-processing files--clean
classifyCleaning of pre-classification tmp filesclassifyMulti coresclassify/pre-processSpeed up kmer countingpre-processEven sized binspre-processOverlapping clusters or tricks for higher accuracy
Author: Sylvain Riondet, PhD student at the National University of Singapore, School of Computing
Email: sylvainriondet@gmail.com
Lab: Genome Institute of Singapore / National University of Singapore
Supervisors: Niranjan Nagarajan & Martin Henz
Thanks for your support and supervision all along my PhD and this project: Martin Henz, Chenhao Li, Rafael Peres, D. Bertrand and the whole MTMS lab