Implementation of Generalized R-squared (refer to the first reference)
For num.noise noise levels and num.type function types, repeat n1 times under null hypothesis to calculate the rejection cutoffs, then repeat n2 times under alternative hypothesis to calculate the statistical power.
- GNU Scientific Library (GSL)
- openmp (speed up the program)
- Rcpp (speed up the original R program)
Make sure you have GSL installed, then run the Makefile
makeyou would get the shared library .so
Then in the R console, run the R code in g2_with_extern.R
Use the following commands to plot like in the original paper. The following code can be directly downloaded from plot.R
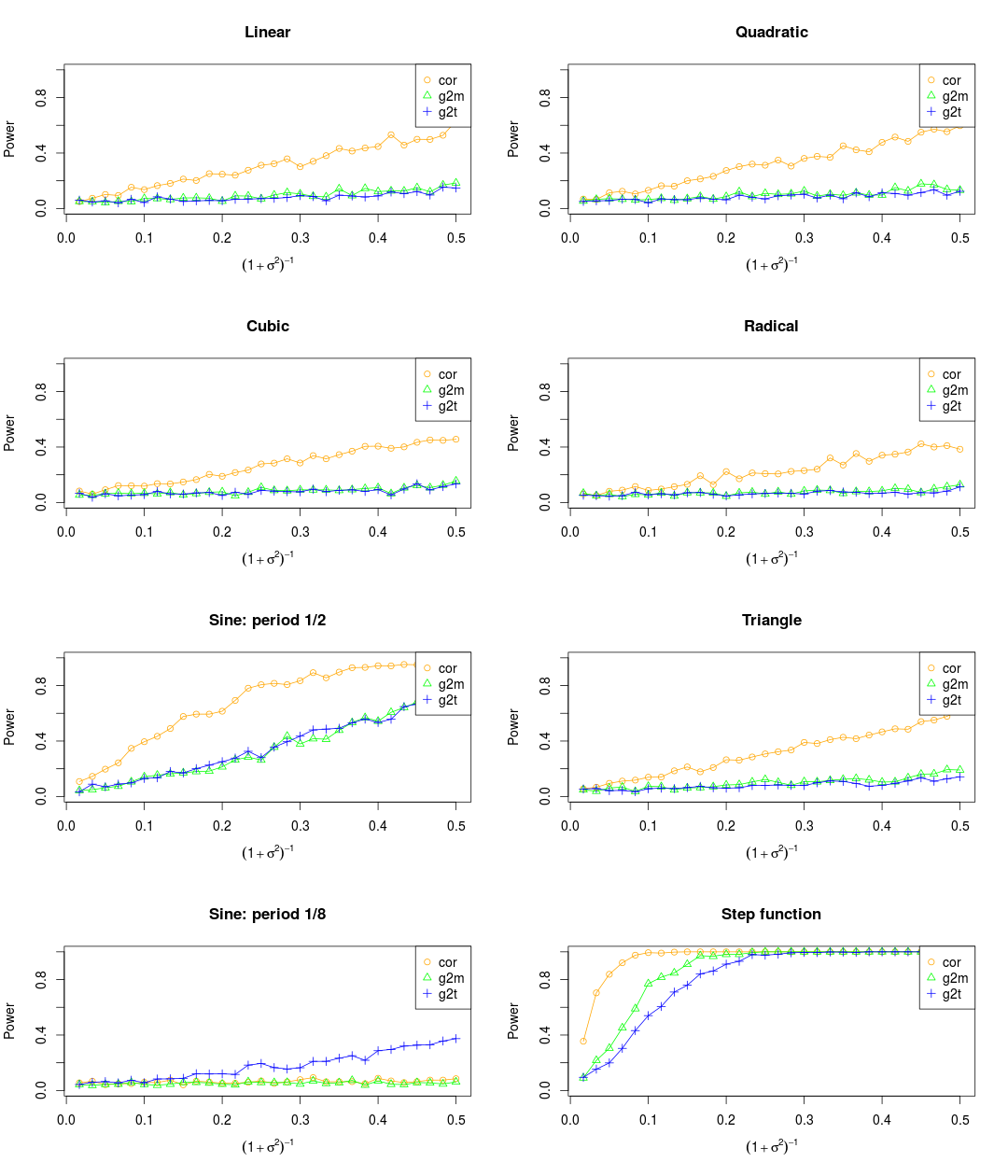
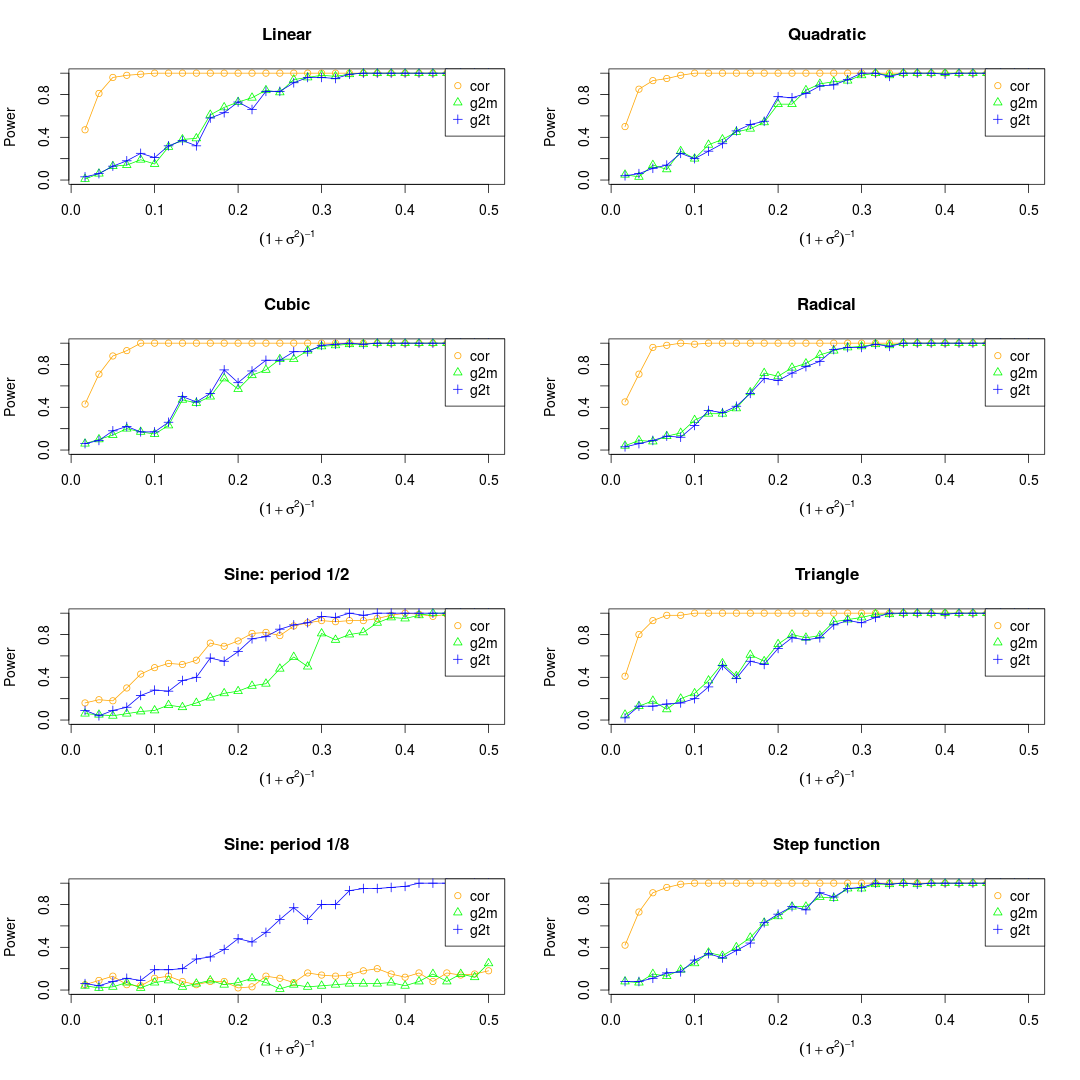
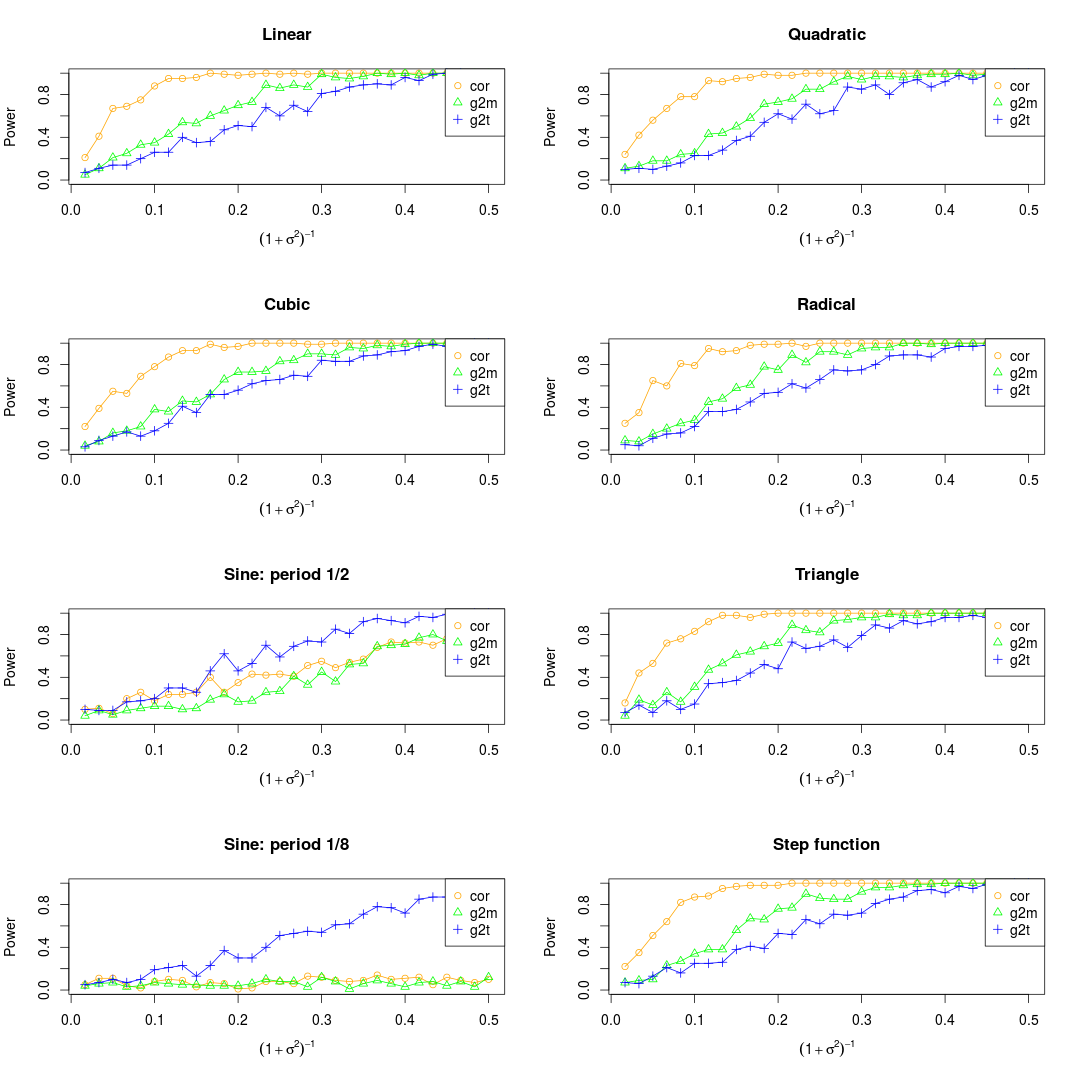
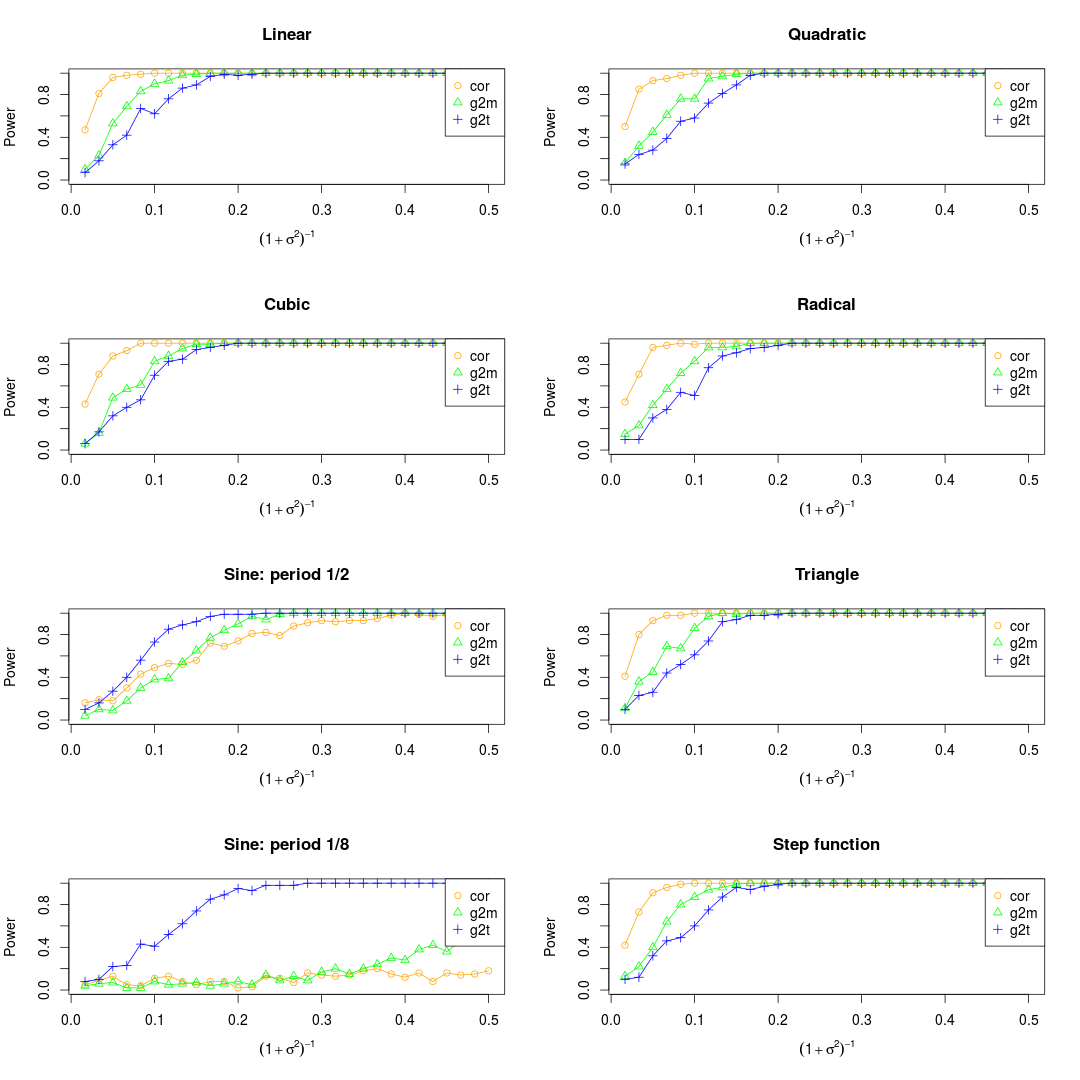
NOTE: There are three different ways to deal with the noise. One is to add the noise to the observations before normalization, another one is to add the noise after normalization, while the third one is to combine them. As far as I'm concerned, I prefer to the third ways.
Set the maximum of x as 0.2
Set the maximum of x as 0.5
The code is as follows:
library(Rcpp)
N = 1000
Rcpp::sourceCpp("g20.cpp")
x = 1:N
gap = array(NA, c(2, 20))
for (i in 1:20)
{
y = sin(1:N) + rnorm(N, sd = i)
df = data.frame(x=x, y=y)
res1 = g2cpp(df)
res2 = g2(x, y)
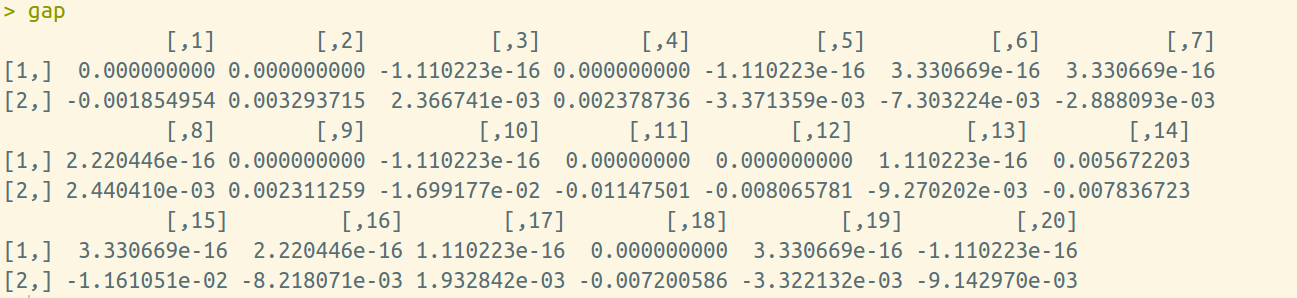
gap[1, i] = res1$g2m - res2$g2m
gap[2, i] = res1$g2t - res2$g2t
}We would get the following results
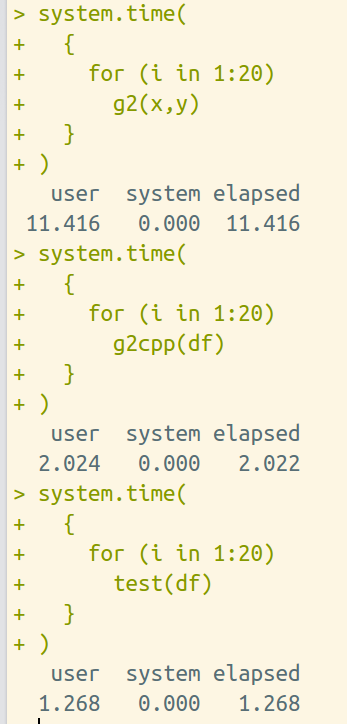
Our original goal is to speed up the program by Rcpp. The following results demonstrate the Rcpp program works!
- Wang, X., Jiang, B., & Liu, J. S. (2017). Generalized R-squared for detecting dependence. Biometrika, 104(1), 129-139.
- Simon N. and Tibshirani R. :http://statweb.stanford.edu/~tibs/reshef/script.R