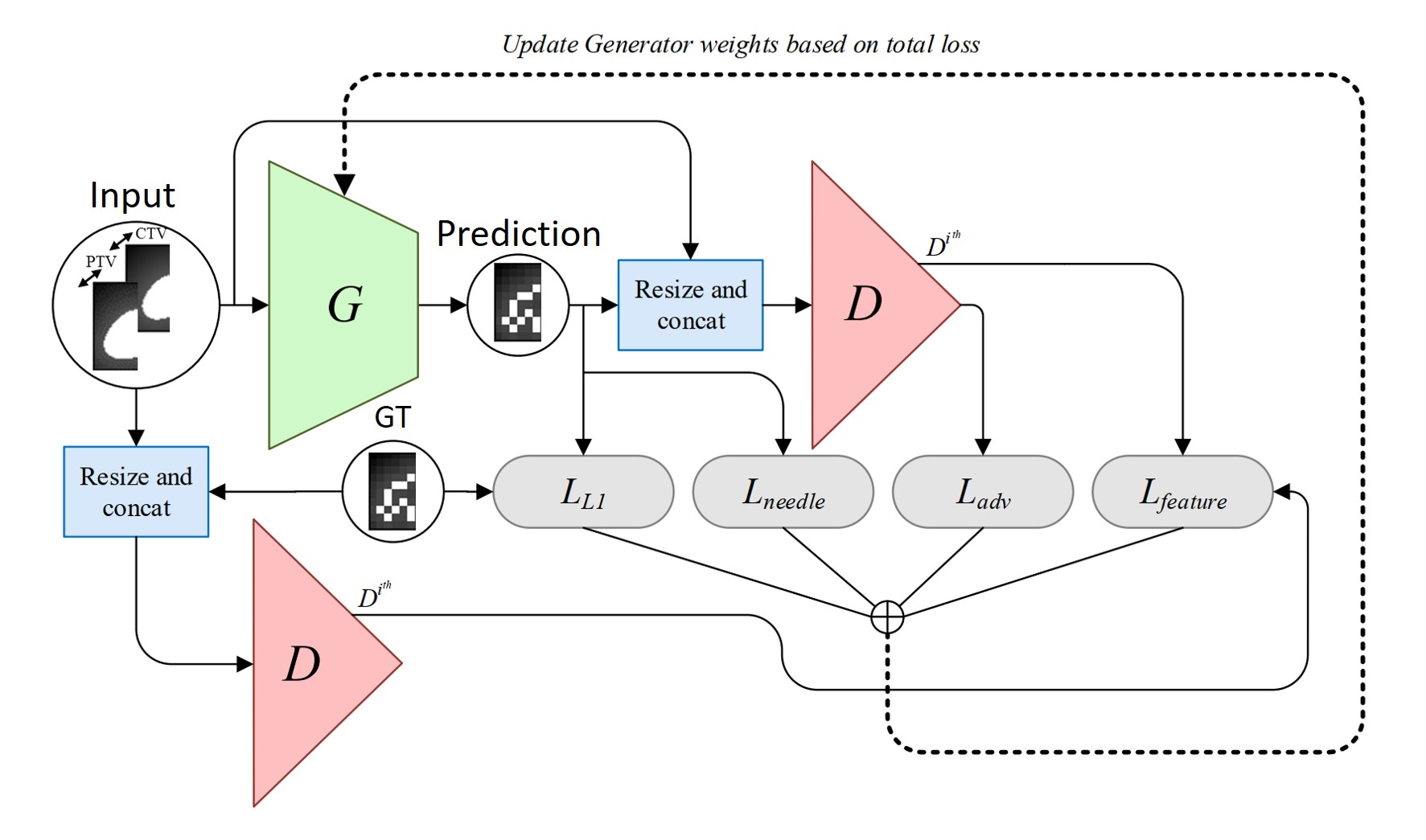
Keras/Tensorflow implementation of 3D pix2pix for automating treatment planning for low-dose-rate prostate brachytherapy. This work has been published in International Journal of Computer Assisted Radiology and Surgery (2021) and was presented in IPCAI 2021.
Python 3.6
Tensorflow: 2.0.0
Keras: 2.3.1
Recreate conda environment as follows:
conda env create -f environment.yml
Or if you are using Docker:
docker pull tazleef/tf2.0.0-cv-keras2.3.1-imgaug:latest
Due to privacy policy, we are unable to share our clinical dataset. However, we have included a few sample cases for reference. Format your centre's dataset in the same way and set the filepath and training parameters in train.py.
To train the model, run train.py.
This code can be used for other 3D image to image translation task by modifying the network architectures according to the data dimensions.
A follow-up of this work can be found here.
@article{aleef2021centre,
title={Centre-specific autonomous treatment plans for prostate brachytherapy using cGANs},
author={Aleef, Tajwar Abrar and Spadinger, Ingrid T and Peacock, Michael D and Salcudean, Septimiu E and Mahdavi, S Sara},
journal={International Journal of Computer Assisted Radiology and Surgery},
pages={1--10},
year={2021},
publisher={Springer}
}
If you face any problem using this code then please create an issue in this repository or contact me at tajwaraleef@ece.ubc.ca
The 3D Resnet code is based on https://github.com/JihongJu/keras-resnet3d
MIT