This repository includes the following spm apps:
- BIDS: for dicom to BIDS conversion and for parsing a BIDS folder. BIDS is the standard for neuroimaging data organization: http://bids.neuroimaging.io
- Boutiques: for running any dockerized neuroimaging tool (e.g. FSL, Freesurfer, ANTs, ...). Boutiques is the standard describing these tools https://boutiques.github.io/
- Quality Control: includes a module for exporting nifti results as an interactive Html page and an advanced Nifti viewer with ROI tools (for drawing a mask).
- Install spm12 if you don't have it: https://www.fil.ion.ucl.ac.uk/spm/software/spm12/
- [R2014b or later] Copy-Paste in your Matlab command line:
% CHANGE DIRECTORY to default Matlab path
if isempty(userpath), userpath('reset'); end
cd(userpath)
% DOWNLOAD spm-apps
websave('spm12-apps.zip','https://github.com/tanguyduval/spm12-apps/archive/master.zip')
unzip spm12-apps.zip
% ADD to Matlab path
addpath(fullfile('spm12-apps-master','BIDS'))
addpath(fullfile('spm12-apps-master','Boutiques'))
addpath(fullfile('spm12-apps-master','QualityControl'))
savepath
% RUN spm Batch Editor
cfg_util('initcfg')
cfg_ui[R2014a or older] In spm Batch Editor:
File-->Add Application-->select cfg_mlbatch_appcfg.m in each folder
create a participant BIDS pipeline in just a minute
- download an example BIDS dataset: https://openneuro.org/datasets/ds001378/versions/00003
- open the matlabbatch using command
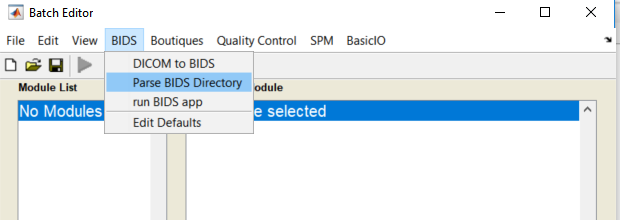
cfg_ui - Add a new module Parse BIDS
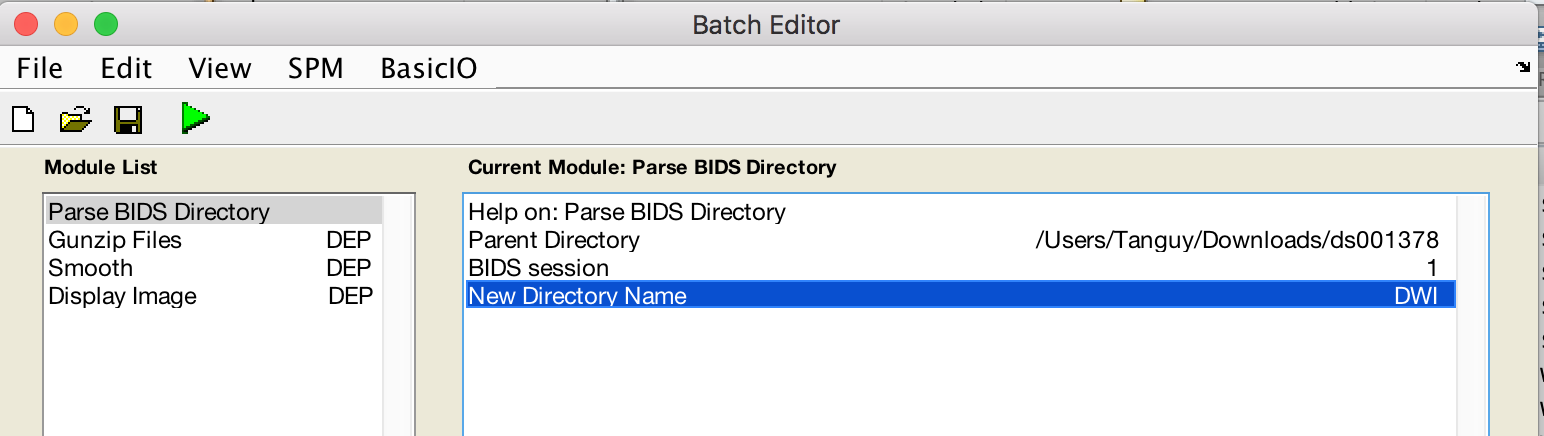
- Fill module as follows
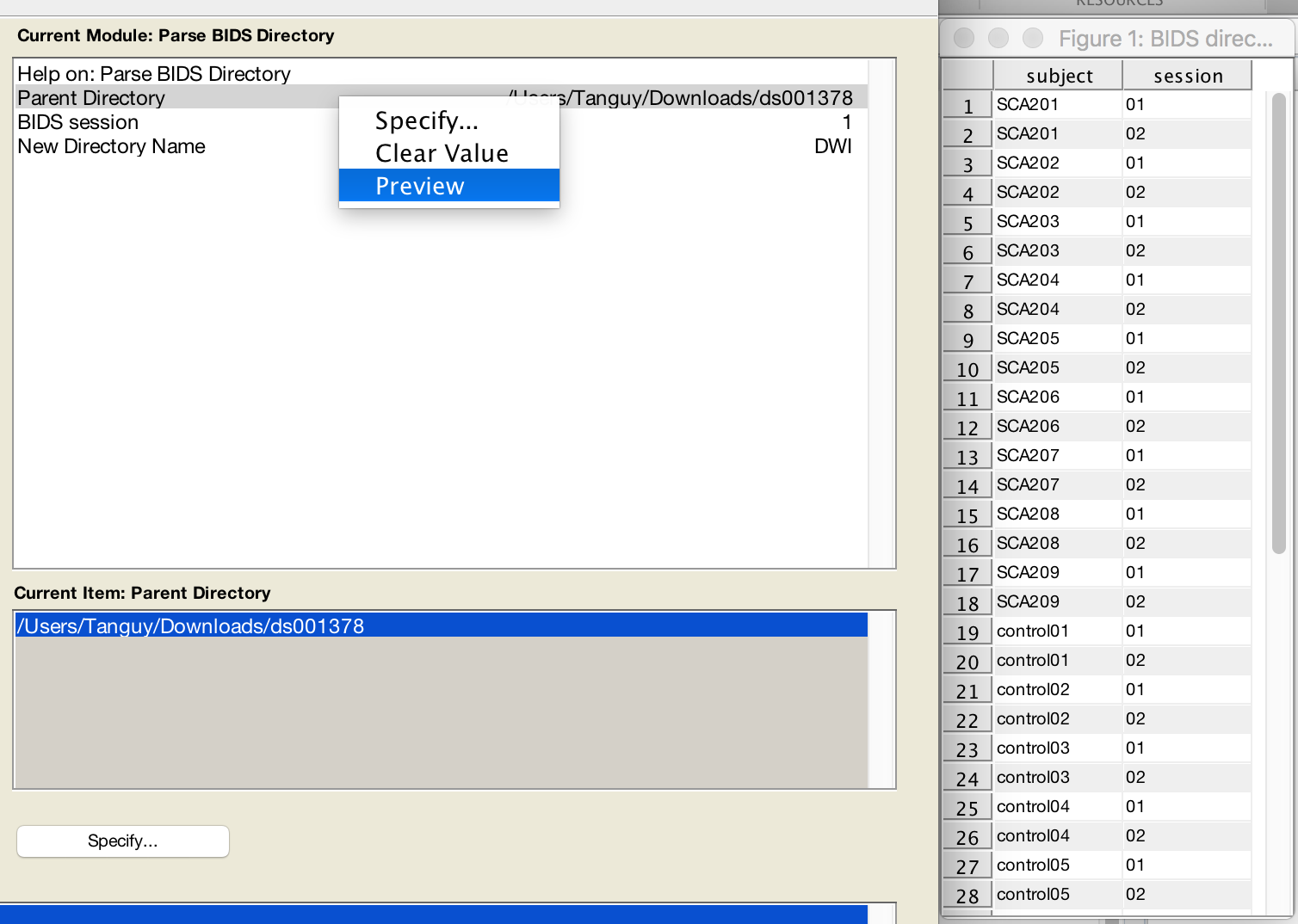
- Preview your BIDS dataset
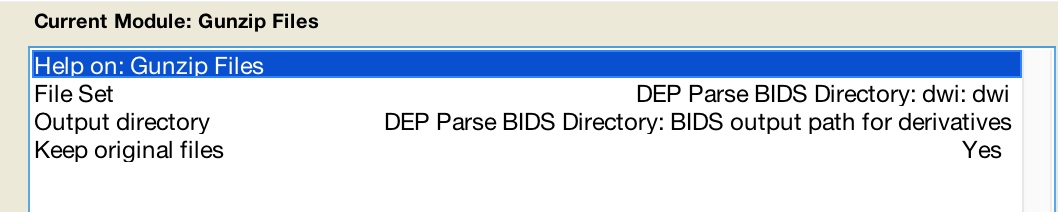
- add a new module BasicIO>FileOperation>Gunzip files
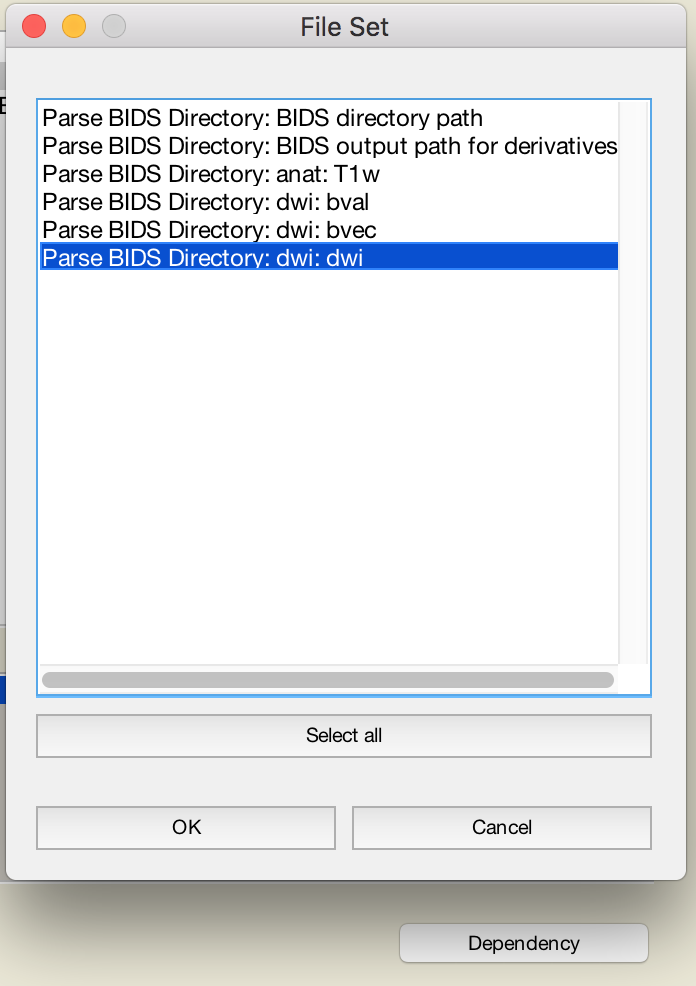
- Use the dependency button and select one modality (e.g. dwi) and fill as follows
- Add additional modules or just click the run icon. Subject 1 session 1 will be processed.
Results will be saved intobidsfolder/derivatives/matlabbatch/sub-NAME/ses-SESSION/DWI/ - Click on the run for all subjects icon (double play icon) to loop across all subjects/sessions
- Save your single participant pipeline using the save icon into a m-file
- Share your pipeline.
(run FSL, ANTS, Freesurfer, ... or your own app)
- install and run docker (https://www.docker.com/get-started). highly recommanded. Note: On windows 8 and older, install docker toolbox: https://docs.docker.com/toolbox/toolbox_install_windows/
- Download a 4D image https://openneuro.org/crn/datasets/ds001378/snapshots/00003/files/sub-control01:ses-01:dwi:sub-control01_ses-01_dwi.nii.gz
- open the Batch Editor using command
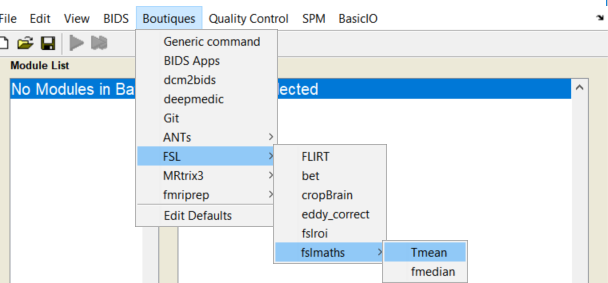
cfg_ui - Use a fslmaths>Tmean module to average across time:
-
Add a new Generic module
- Fill the module as follows
Use Docker: docker images with preinstalled neuroimaging softwares can be found on https://hub.docker.com/
bids/mrtrix3-connectomeincludes ANTS MRtrix3 and FSL docker images will be automatically downloaded and inputs/output folders mounted
if you choose "NO", command will be run locally
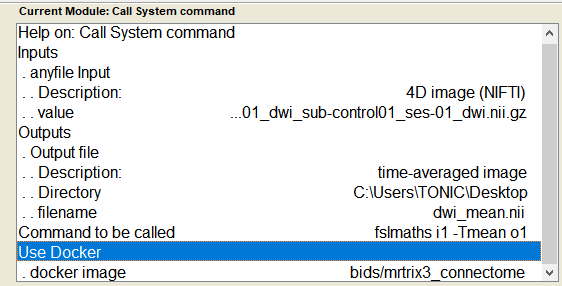
- RUN the command using the green play button
- Save your preset module as a Boutiques descriptor https://boutiques.github.io
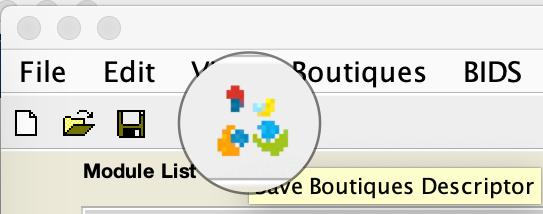
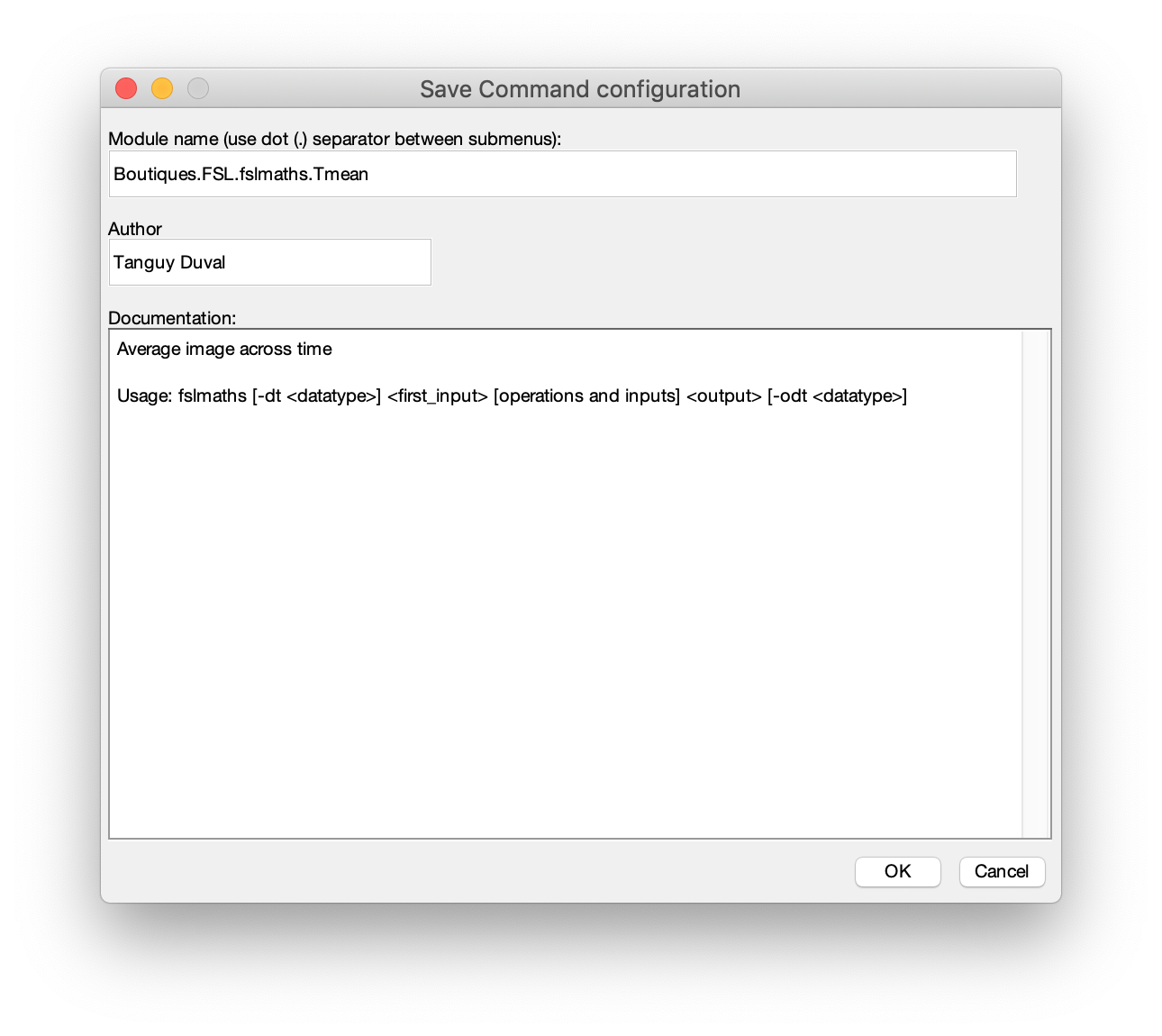
- Reload modules
- Fill the module as follows
cfg_util('initcfg')
cfg_ui- Use your preset module in an other pipeline
- Your preset is saved under
spm-apps\Boutiques\Boutiques