MARGARET is a computational tool for Trajectory Inference i.e. for inferring trajectories underlying biological developmental processes like cell-differentiation. More specifically the methodology can be sub-divided into three major steps:
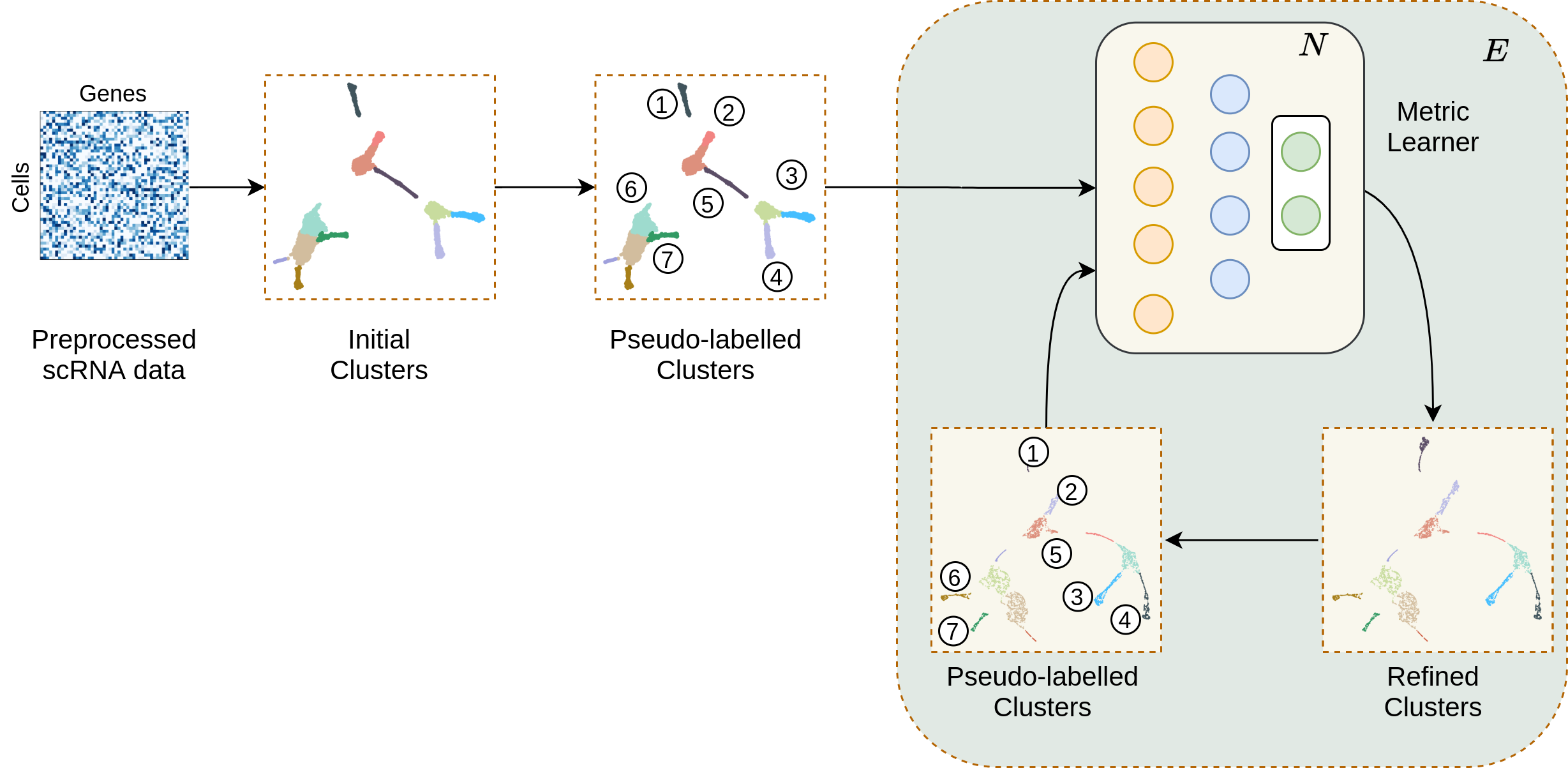
MARGARET utilizes a deep unsupervised metric learning-based approach for inferring low-dimensional cellular embeddings and cluster assignments at a single-cell (sc) level from omics-type data (e.g. transcriptomics (scRNA-seq), proteomics (CITE-seq), among many others)
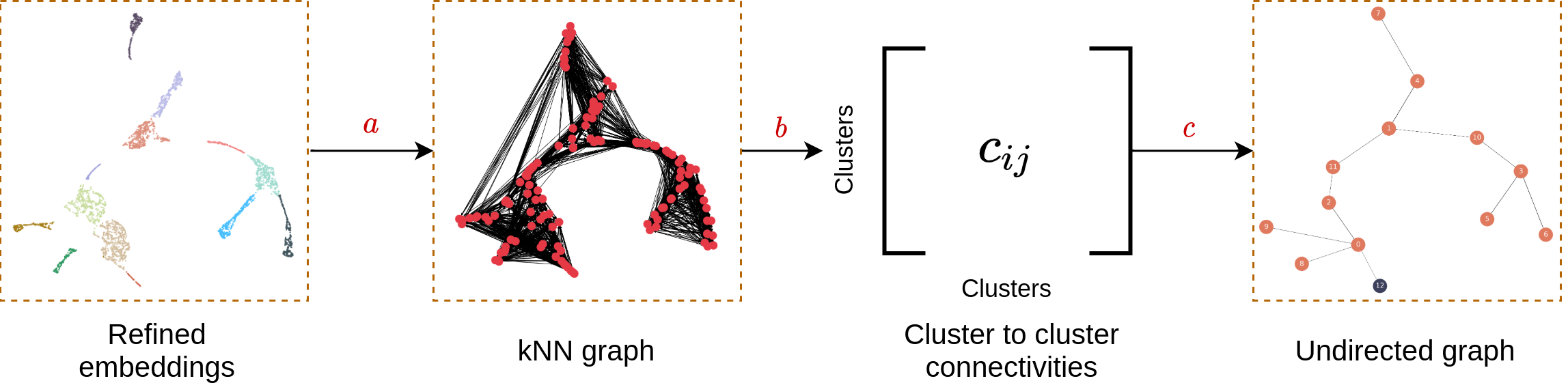
MARGARET employs a novel measure of connectivity to assess connectivity between the inferred clusters in the first step and constructs a cluster-level undirected graph to represent a trajectory topology as shown in the following workflow.
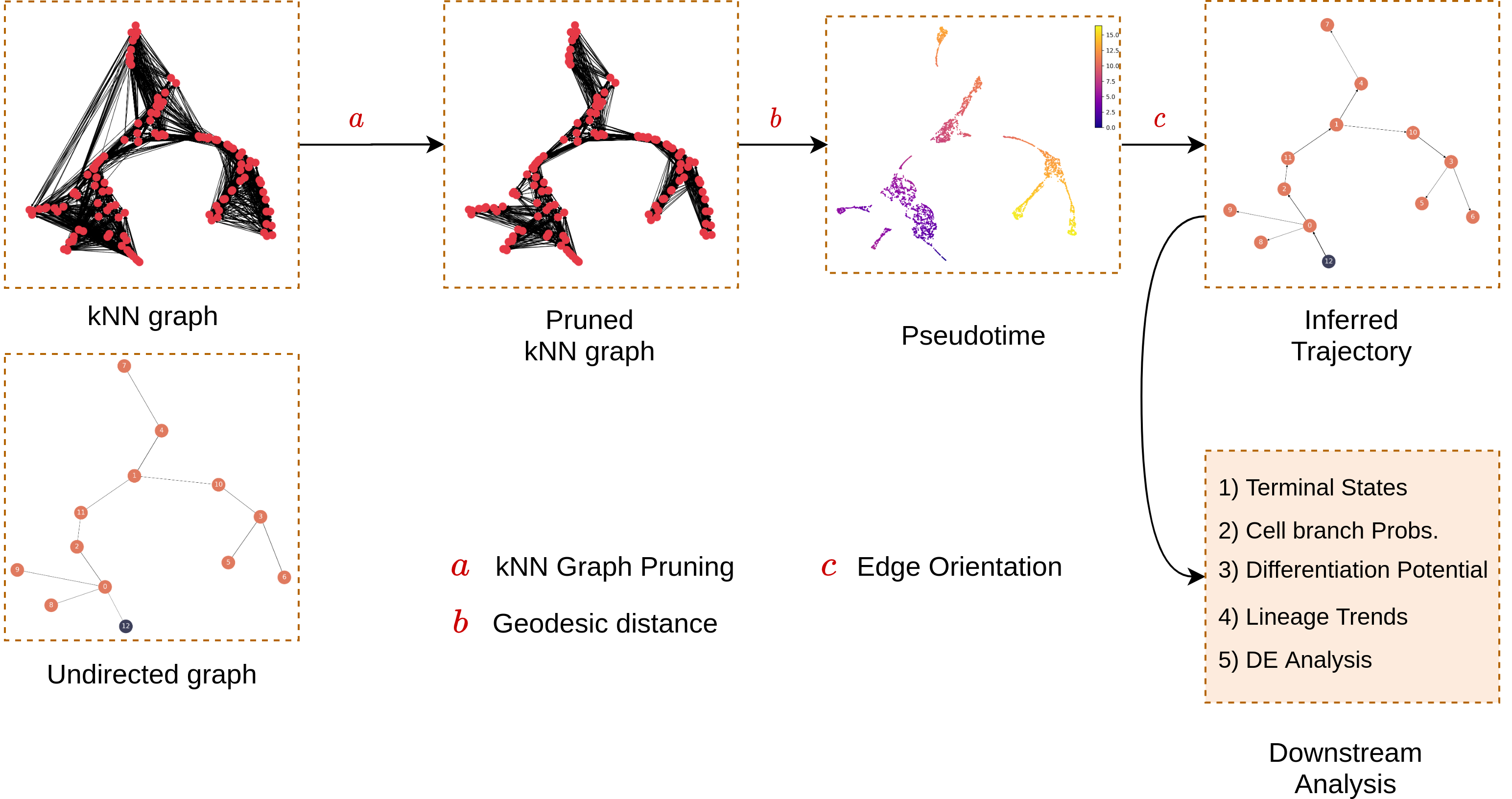
MARGARET then contructs a kNN graph between all cells (on a single-cell level) and prunes it with reference to the undirected graph computed previously. With a starting cell specified, MARGARET then assigns a pseudotime to each cell in the pruned kNN graph denoting the position of this cell in the underlying trajectory.
Given the pseudotime for each cell and the undirected graph inferred in Step 2, MARGARET directs the edges in the undirected graph which represents the final underlying trajectory.
Different inferred components (like the trajectory, cell embeddings and clusters) can then be utilized for downstream tasks like Terminal state detection and cell-fate prediction.
For more details, refer to our manuscript "Inference of cell state transitions and cell fate plasticity from single-cell with MARGARET" at https://www.biorxiv.org/content/10.1101/2021.10.22.465455v1
A stable pip installable release for MARGARET will be made available shortly. For now, we recommend users to directly clone our stable main branch and set margaret as the working directory. All package dependencies for using MARGARET can be setup as follows:
- Install
pipenvby runningpip install pipenv - With
margaretas the working directory runpipenv installto setup all required dependencies.
Running pipenv for the first time will auto-create a project wide virtual environment :)
MARGARET can be used for:
- Computing low-dimensional cell embeddings from the single-cell data.
- Automatically inferring clusters from the data.
- Inferring a cluster-level trajectory underlying the data
- Pseudotime-inference
- Automated terminal state detection
- Quantifying cell-fate plasticity at a cell-level.
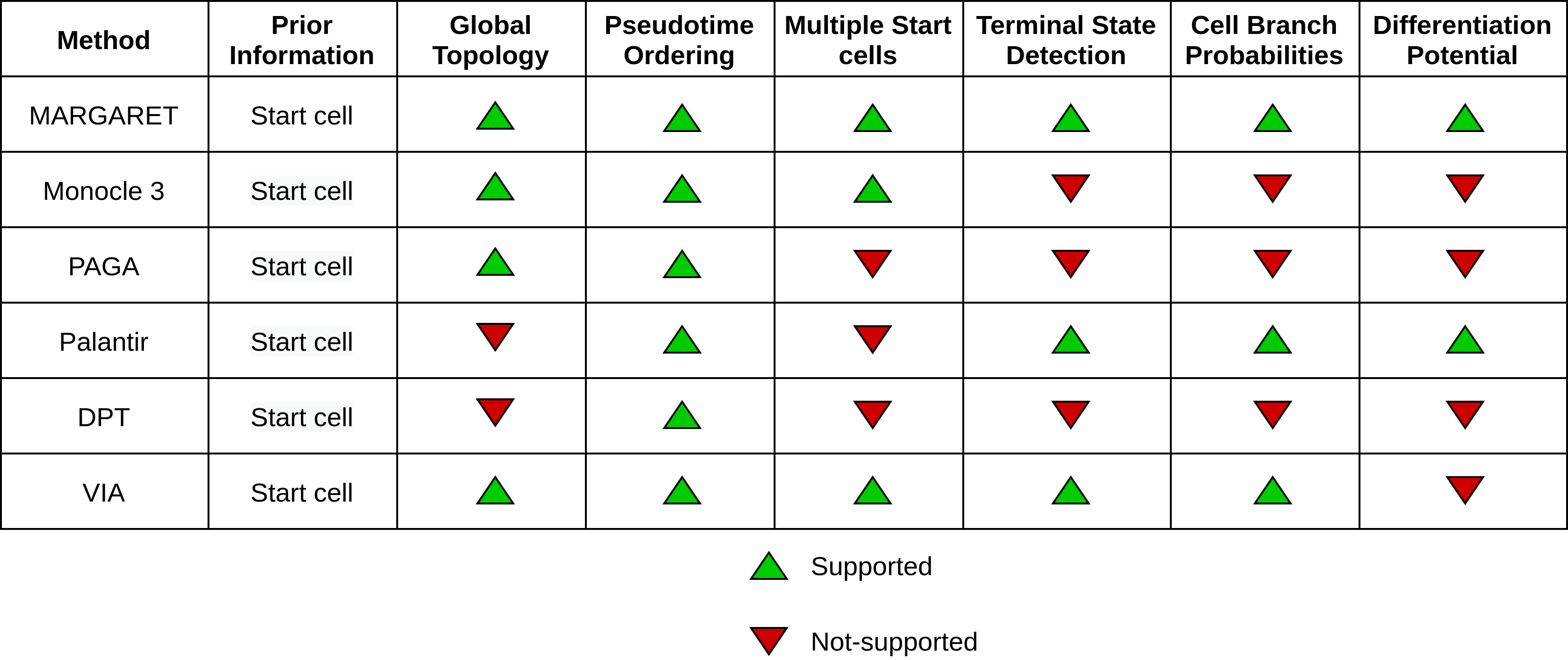
MARGARET supports several features that prior works in Trajectory Inference do not support. Moreover, on a diverse benchmark MARGARET outperforms prior methods in TI. We encourage interested readers to refer to our manuscript for more details on comparisons.
Check out the following Colab notebooks to get a flavor of a typical workflow in Trajectory Inference and MARGARETs capabilities:
- MARGARET applied to simulated datasets
- MARGARET applied to scRNA-seq data for early human hematopoiesis
- MARGARET applied to scRNA-seq data for early embryogenesis
In case of any bug reports, enhancement requests, general questions, and other contributions, please create an issue. For more substantial contributions, please fork this repo, push your changes to your fork, and submit a pull request with a good commit message.
Kushagra Pandey, Hamim Zafar