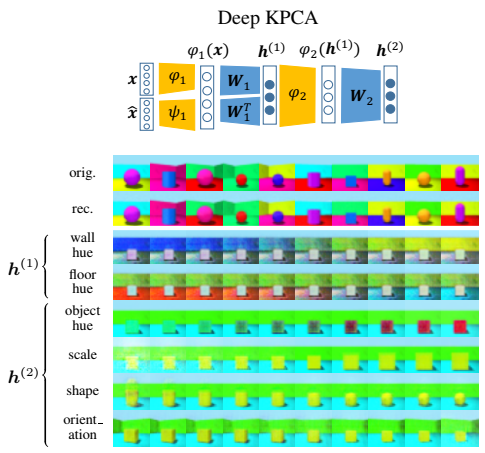
Deep KPCA is a deep kernel method to learn efficient and disentangled data representations in multiple levels, with forward and backward dependency across levels.
Principal Component Analysis (PCA) and its nonlinear extension Kernel PCA (KPCA) are widely used across science and industry for data analysis and dimensionality reduction. Modern deep learning tools have achieved great empirical success, but a framework for deep principal component analysis is still lacking. Here we develop a deep kernel PCA methodology (DKPCA) to extract multiple levels of the most informative components of the data. Our scheme can effectively identify new hierarchical variables, called deep principal components, capturing the main characteristics of high-dimensional data through a simple and interpretable numerical optimization. We couple the principal components of multiple KPCA levels, theoretically showing that DKPCA creates both forward and backward dependency across levels, which has not been explored in kernel methods and yet is crucial to extract more informative features. Various experimental evaluations on multiple data types show that DKPCA finds more efficient and disentangled representations with higher explained variance in fewer principal components, compared to the shallow KPCA. We demonstrate that our method allows for effective hierarchical data exploration, with the ability to separate the key generative factors of the input data both for large datasets and when few training samples are available. Overall, DKPCA can facilitate the extraction of useful patterns from high-dimensional data by learning more informative features organized in different levels, giving diversified aspects to explore the variation factors in the data, while maintaining a simple mathematical formulation.
- Training and evaluation of a DKPCA model is done in
train*.pyfrom themain.- Training a two-level DKPCA with nonlinear first level and linear second level is done in
train_lin.py. - Training DKPCA with RBF kernel in both two levels is done in
train_nl.py. The four RBF levels experiments are done withtrain_nl4.py. - Training DKPCA with explicit feature maps is done in
train_cnn.py.
- Training a two-level DKPCA with nonlinear first level and linear second level is done in
- Disentanglement evaluation is done in
gen.py(latent traversal plots) and ineval_irs.py(quantitative evaluation). - Available kernels are in
kernels.py. - Available explicit feature maps (both encoder and decoder) are in
train_cnn.py. - Employed datasets are in
dataloader.py.
First, navigate to the unzipped directory and install required python packages with the provided requirements.txt file. This is explained in the following section.
Run the following in terminal. This will create a conda environment named rkm_env.
conda create --name rkm_env python=3.10
Activate the conda environment with the command conda activate rkm_env. To install the required dependencies, run:
pip install -r requirements.txt
Activate the conda environment by conda activate rkm_env and run one of the following commands, for example:
python train_lin.py dataset=multivariatenormal levels=j2_linear levels.j1.s=2 levels.j2.s=2
python train_nl.py dataset=3dshapes dataset.N=100 levels=j2_rbf
python train_cnn.py dataset=3dshapes dataset.N=1000 levels=j2_cnn_lin optimizer=geoopt optimizer.maxepochs=10
The configuration is done using YAML files with hydra. Available configurations are in the configs directory. You may edit the provided YAML files or write your own files. Alternatively, you can change configurations directly from the command line, e.g,.:
python train_lin.py levels.j2.s=5
This can be done in dataloader.py.
To add a new dataset, define a new get_dataset_dataloader function returning the pytorch dataloader. Then, add that dataset to the get_dataloader_helper function.
This can be done in kernels.py.
To add a new kernel, define a new pytorch module, similar to, e.g., GaussianKernelTorch. Then, add that kernel to the kernel_factory function.
To run DKPCA with two RBF levels on the synthetic dataset Synth 2 with 30 components in each level, open a terminal in the main directory and run
cd <maindirectoryname>
python train_nl.py dataset=complex6 dataset.N=100 levels=j2_rbf levels.j1.s=30 levels.j2.s=30 optimizer=pg optimizer.maxepochs=5000 levels.j2.eta=1 optimizer.torch.lr=0.01
The training and the evaluation are shown on screen. The model is saved in $HOME/out/deepkpca for future use.
To run DKPCA on the real-world datasets MNIST/3dshapes/Cars3d/SmallNORB, you may use a similar command structure. For example, to train DKPCA with two RBF levels on a subset of 3dshapes
cd <maindirectoryname>
python train_nl.py dataset=3dshapes dataset.N=50 levels=j2_rbf levels.j1.s=30 levels.j1.kernel.args.sigma2=100 levels.j2.s=30 levels.j2.kernel.args.sigma2=1 optimizer=pg optimizer.torch.lr=0.01 optimizer.maxepochs=5000
The employed benchmark datasets are downloaded in $HOME/data. Their availability is described in the dataloader.py file.
In this section, we explain how to evaluate the disentanglement performance of DKPCA, both qualitatively with latent traversals and quantitatively with disentanglement metrics. We also describe how to run the compared deep learning methods.
After training a DKPCA model, the model weights are saved in $HOME/out/deepkpca/<model_label>, i.e., a label is associated with every trained model. The traversals along the deep principal components, as well as the reconstructions, are then performed in gen.py from the given pre-trained model label. For instance, if the model label is 3pp8rxvi, the generation is done by running:
python gen.py --model 3pp8rxvi
The generated images are saved in the Traversal_imgs directory.
The arguments of the main also specify the number of steps in the traversals and the variance of the Gaussian distribution from which the new vectors of hidden features are sampled from the latent space.
Computation of the IRS metric is done in eval_irs.py. For instance, if the trained model label is 3pp8rxvi, the quantitative disentanglement evaluation is done by running:
python eval_irs.py --model 3pp8rxvi --n 1000
The command line argument --n specifies the number of samples to use for evaluation.
Before running eval_irs.py, the employed disent library needs to be updated to compute the IRS metric and to handle subsampling with an additional optional available_y argument to the StateSpace.sample_factors method. To do this, copy the files in the provided disent_irs directory to the directory of the disent library. For instance, if conda was installed in your home directory
cp -r disent_irs/* ~/miniconda3/envs/rkm_env/lib/python3.10/site-packages/disent/
The python script eval_irs.py can also be used in the same way to evaluate VAE-based models trained using the disentanglement_lib library for the compared deep learning methods (see next section for details). For that library, Tensorflow is an additional dependency to be installed.
The VAE-based methods beta-VAE, FactorVAE, and beta-TCVAE were trained with the disentanglement_lib library for both the small- and large-scale experiments. For the Cars3D large-scale quantitative comparison and for the latent traversals, the pre-trained models are obtained following disentanglement_lib documentation.
To verify the deep approximation lower and upper bounds numerically, run
python train_lin.py optimizer.maxepochs=10000 levels=j2_rbf_lin dataset.N=10 levels.j1.s=10 levels.j2.s=10 bounds=true
The following options for training a DKPCA model are available:
$ python train_lin.py --help
train_lin is powered by Hydra.
== Configuration groups ==
Compose your configuration from those groups (group=option)
dataset: 3dshapes, cars3d, cholesterol, complex6, diabetes, ionosphere, liver, mnist, multivariatenormal, norb, square, yacht
levels: j2_cnn_lin, j2_linear, j2_rbf, j2_rbf_lin, j4_rbf
model: rkm
optimizer: adam, geoopt, pg
== Config ==
Override anything in the config (foo.bar=value)
seed: 0
saved_model: null
ae_weight: null
bounds: false
dataset:
name: square
'N': 100
mb_size: 64
Ntest: -1
shuffle: false
workers: 0
post_transformations: []
pre_transformations: []
val_perc: 0
std: 0.05
levels:
j1:
lambd: 1.0
eta: 1.0
s: 2
kernel:
name: linear
args: null
j2:
lambd: 1.0
eta: 1.0
s: 2
kernel:
name: linear
args: null
optimizer:
name: pg
torch:
lr: 0.01
init:
name: random
maxepochs: 1000
epsilon: 0.001
beta: 1.0e-07
patience: 1000
model:
name: rkm
svdopt: true
Powered by Hydra (https://hydra.cc)
Use --hydra-help to view Hydra specific help