Implement random forest and BRAF from scratch.
To train a random forest (RF) or Biased Random Forest (BRAF) on the diabetes dataset with k-fold cross validation, and to save ROC and PRC curves use:
python evaluate_forest.py --forest BRAF --k 10 --folds 10 --p_ratio 0.5 --size 100 --print_plot --evaluate_on_train_set --k_fold_cross_val
Arguments:
--forest: BRAF or RF
--k: BRAF argument k, for k-nearest neighbors in cirical data set
--folds: Number of folds for cross-validation
--p_ratio: BRAF argument. Relative ratio of random forests trained on
critical/full data set
--size 100: Number of trees in random forest
--k_fold_cross_val Perform cross validation
--print_plot: Print PRC/ROC curves for each cross validation fold
--evaluate_on_train_set Evaluate final model both train and test set
Data is imbalanced 2:1 between the classes 0/1.
-
Impute by replacing missing values (except for pregnancy feature) by mean of non-missing values
-
Normalize all features to lie between 0 and 1
>>> python evaluate_forest.py --forest BRAF --k 10 --folds 10 --p_ratio 0.5 --size 100 --print_plot --evaluate_on_train_set --k_fold_cross_val
Length of full dataset: 768
Initial train/test split into sets of length: 640 / 128
Perform 10-fold cross validation within train set:
Model: Biased Random Forest. Parameters: size = 100, p = 0.5, k = 10
------------------------------------------------------------------------------------------
| Fold: 0 | Accuracy: 0.75 | Precision: 0.58 | Recall: 0.75 | AUROC: 0.82 | AUPRC: 0.64 |
| Fold: 1 | Accuracy: 0.83 | Precision: 0.75 | Recall: 0.63 | AUROC: 0.86 | AUPRC: 0.72 |
| Fold: 2 | Accuracy: 0.78 | Precision: 0.65 | Recall: 0.77 | AUROC: 0.85 | AUPRC: 0.70 |
| Fold: 3 | Accuracy: 0.78 | Precision: 0.65 | Recall: 0.77 | AUROC: 0.84 | AUPRC: 0.74 |
| Fold: 4 | Accuracy: 0.78 | Precision: 0.83 | Recall: 0.66 | AUROC: 0.81 | AUPRC: 0.82 |
| Fold: 5 | Accuracy: 0.69 | Precision: 0.81 | Recall: 0.52 | AUROC: 0.84 | AUPRC: 0.86 |
| Fold: 6 | Accuracy: 0.73 | Precision: 0.65 | Recall: 0.68 | AUROC: 0.79 | AUPRC: 0.72 |
| Fold: 7 | Accuracy: 0.78 | Precision: 0.64 | Recall: 0.50 | AUROC: 0.77 | AUPRC: 0.62 |
| Fold: 8 | Accuracy: 0.80 | Precision: 0.56 | Recall: 0.67 | AUROC: 0.82 | AUPRC: 0.68 |
| Fold: 9 | Accuracy: 0.72 | Precision: 0.64 | Recall: 0.33 | AUROC: 0.83 | AUPRC: 0.58 |
------------------------------------------------------------------------------------------
Averages +/- Standard Error
---------------------------
Accuracy: 0.76 +/- 0.01
Precision: 0.68 +/- 0.03
Recall: 0.63 +/- 0.04
AUROC: 0.82 +/- 0.01
AUPRC: 0.71 +/- 0.03
Train model on full train set.
Model evaluation on train set:
| Fold: _final_train | Accuracy: 0.98 | Precision: 0.99 | Recall: 0.96 | AUROC: 1.00 | AUPRC: 1.00 |
Model evaluation on test set:
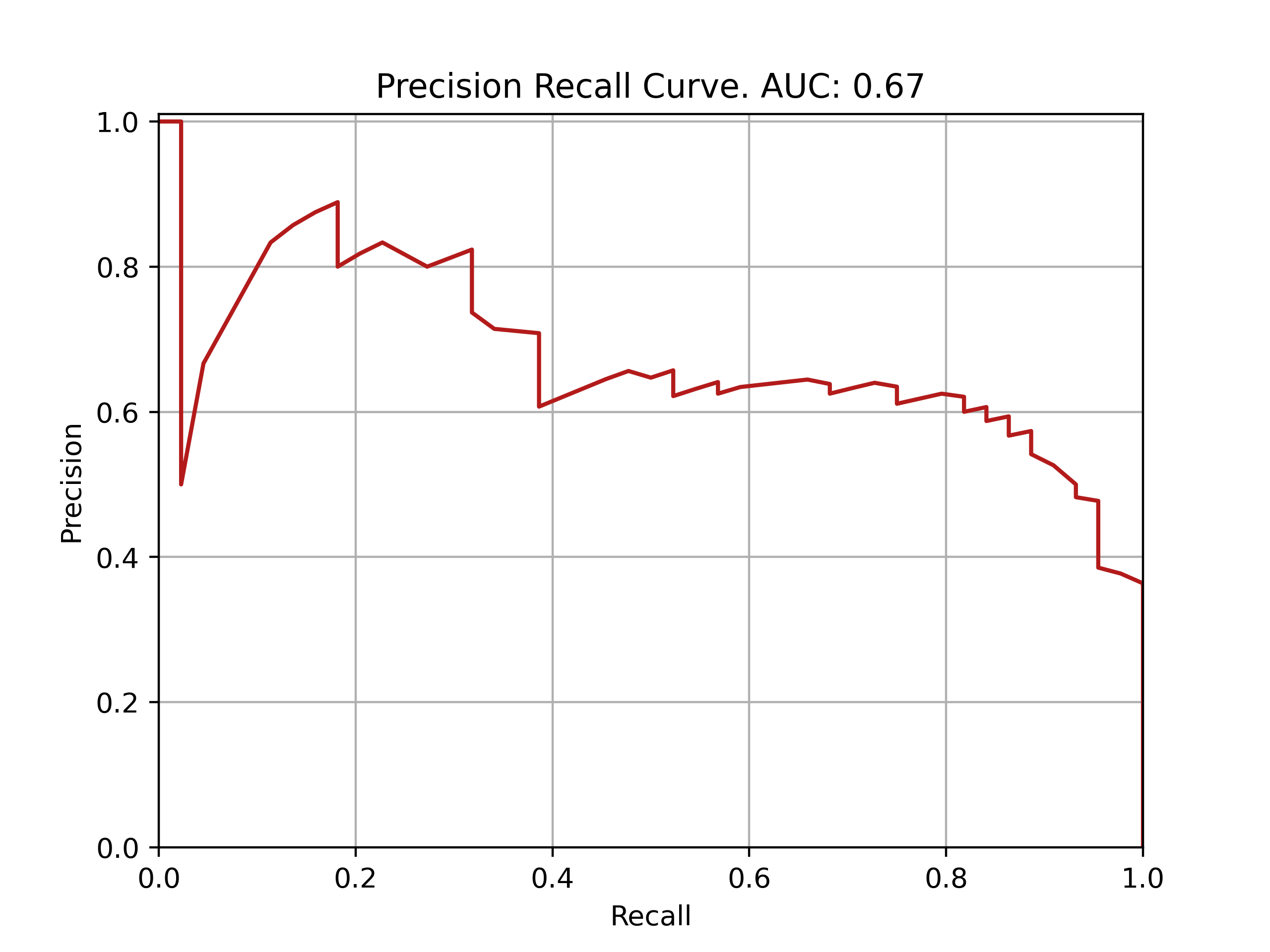
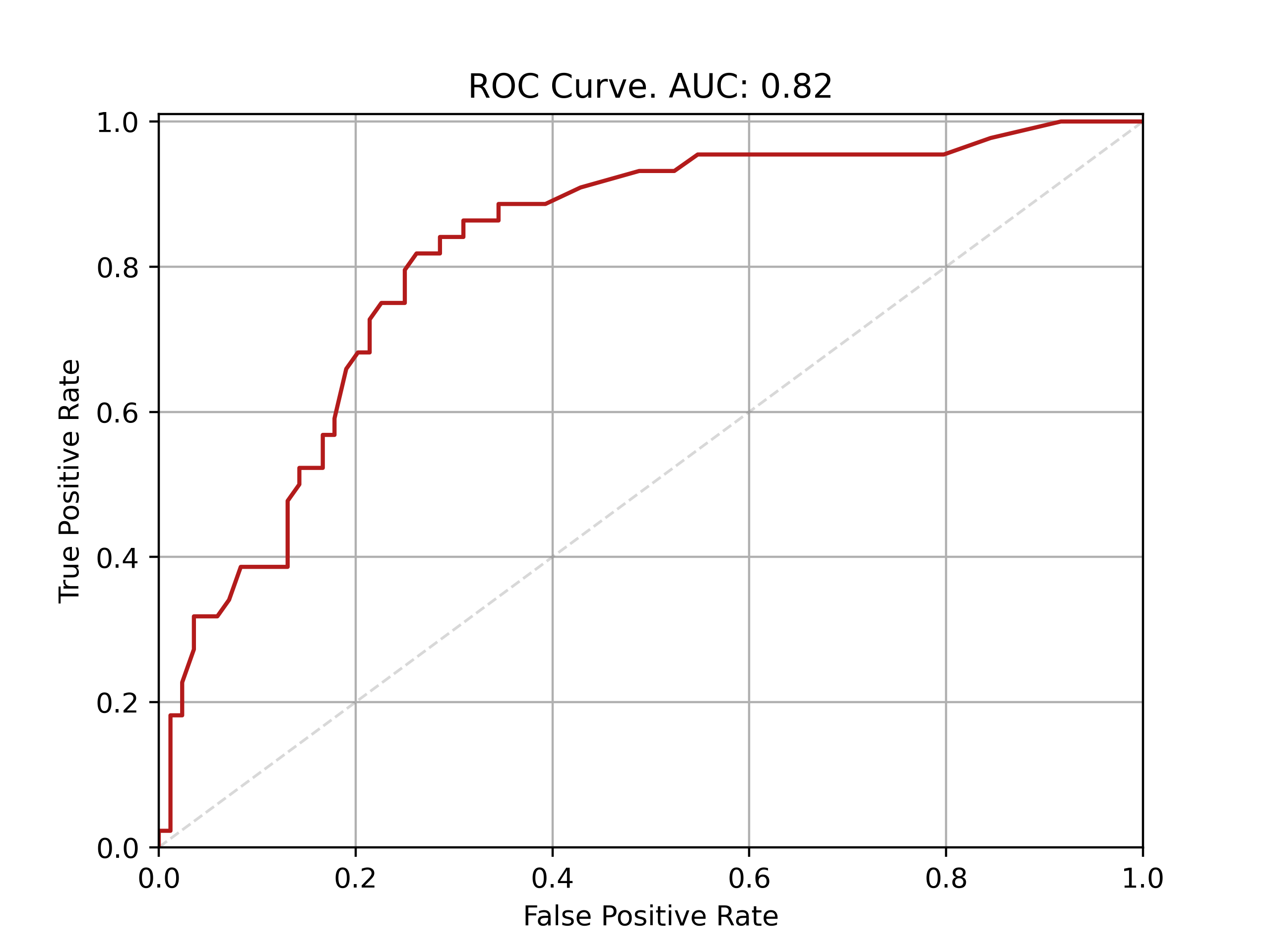
| Fold: _final_test | Accuracy: 0.76 | Precision: 0.64 | Recall: 0.68 | AUROC: 0.82 | AUPRC: 0.67 |
ROC and PRC curves for final model evaluation on test set:
>>> python evaluate_forest.py --forest RF --size 100 --print_plot --evaluate_on_train_set --k_fold_cross_val
Length of full dataset: 768
Initial train/test split into sets of length: 640 / 128
Perform 10-fold cross validation within train set:
Model: Plain Random Forest. Parameters: size = 100
------------------------------------------------------------------------------------------
| Fold: 0 | Accuracy: 0.81 | Precision: 0.70 | Recall: 0.70 | AUROC: 0.90 | AUPRC: 0.81 |
| Fold: 1 | Accuracy: 0.73 | Precision: 0.64 | Recall: 0.43 | AUROC: 0.80 | AUPRC: 0.61 |
| Fold: 2 | Accuracy: 0.81 | Precision: 0.71 | Recall: 0.77 | AUROC: 0.84 | AUPRC: 0.77 |
| Fold: 3 | Accuracy: 0.77 | Precision: 0.76 | Recall: 0.54 | AUROC: 0.83 | AUPRC: 0.77 |
| Fold: 4 | Accuracy: 0.69 | Precision: 0.62 | Recall: 0.42 | AUROC: 0.74 | AUPRC: 0.65 |
| Fold: 5 | Accuracy: 0.80 | Precision: 0.68 | Recall: 0.77 | AUROC: 0.89 | AUPRC: 0.75 |
| Fold: 6 | Accuracy: 0.69 | Precision: 0.52 | Recall: 0.52 | AUROC: 0.76 | AUPRC: 0.56 |
| Fold: 7 | Accuracy: 0.81 | Precision: 0.78 | Recall: 0.64 | AUROC: 0.83 | AUPRC: 0.72 |
| Fold: 8 | Accuracy: 0.73 | Precision: 0.65 | Recall: 0.57 | AUROC: 0.77 | AUPRC: 0.69 |
| Fold: 9 | Accuracy: 0.84 | Precision: 0.80 | Recall: 0.73 | AUROC: 0.84 | AUPRC: 0.69 |
------------------------------------------------------------------------------------------
Averages +/- Standard Error
---------------------------
Accuracy: 0.77 +/- 0.02
Precision: 0.69 +/- 0.02
Recall: 0.61 +/- 0.04
AUROC: 0.82 +/- 0.02
AUPRC: 0.70 +/- 0.02
Train model on full train set.
Model evaluation on train set:
| Fold: _final_train | Accuracy: 0.98 | Precision: 0.98 | Recall: 0.98 | AUROC: 1.00 | AUPRC: 1.00 |
Model evaluation on test set:
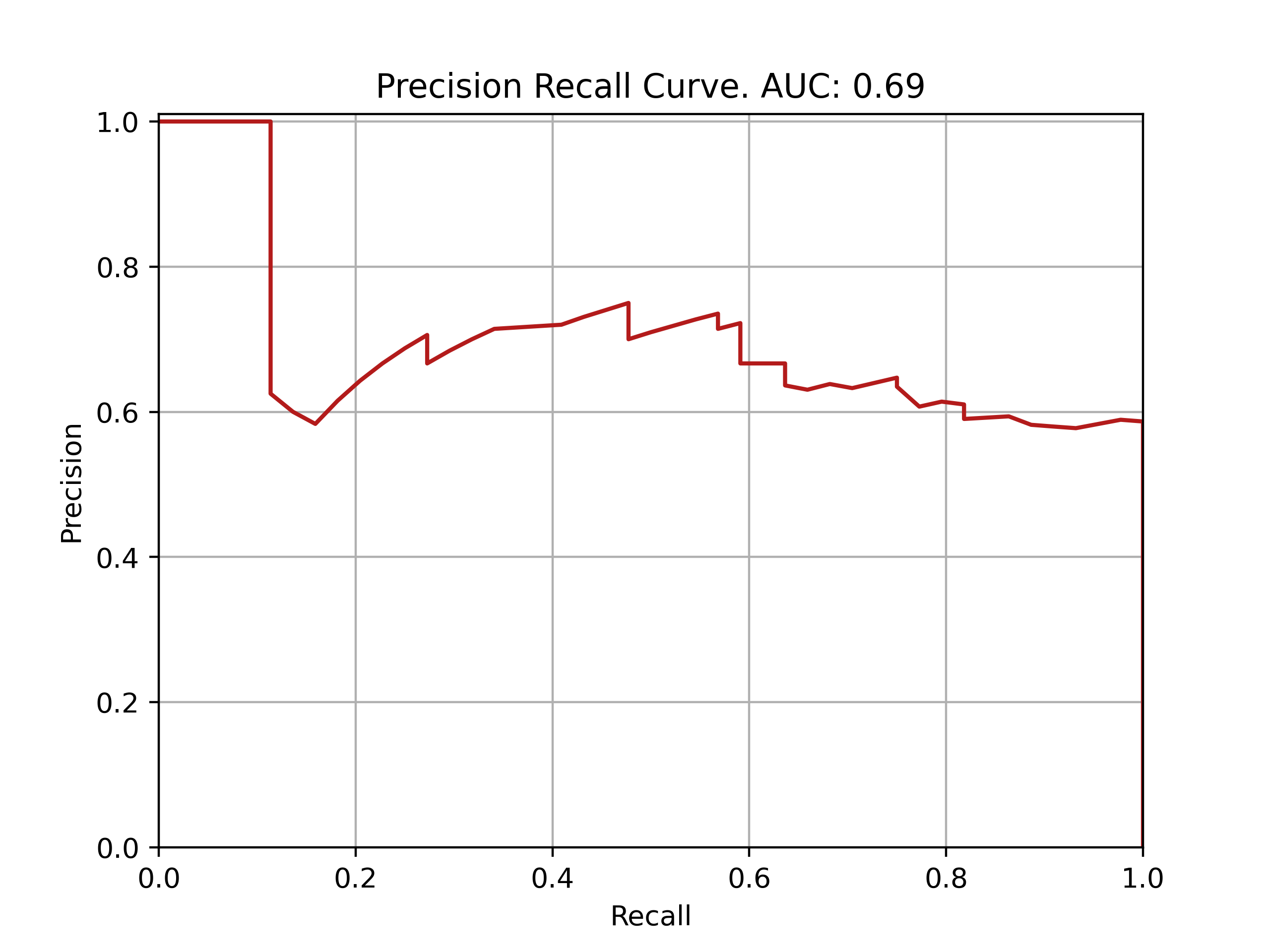
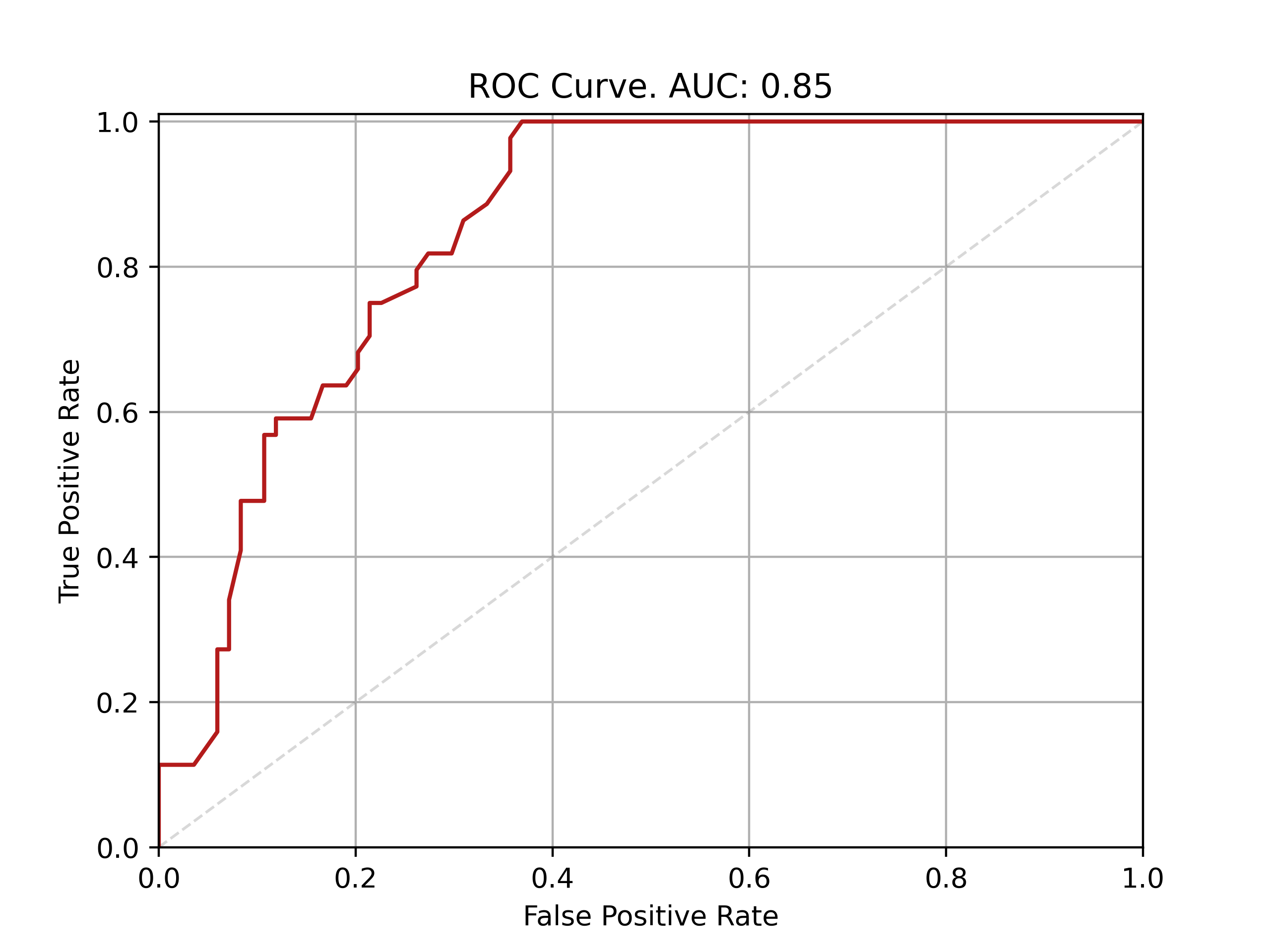
| Fold: _final_test | Accuracy: 0.77 | Precision: 0.73 | Recall: 0.55 | AUROC: 0.85 | AUPRC: 0.69 |
ROC and PRC curves for final model evaluation on test set:
References: https://ieeexplore.ieee.org/document/8541100
- The implementation is in plain Python and NumPy would speed it up.
- The "critical" dataset in BRAF contains almost the entire dataset with the default parameters, which inhibits potential improvements.
- Stratified train/test split might help