Title: A tutorial on interpretable machine learning algorithms for understanding factors related to childhood autism
Abstract: machine learning is a research area in computer science which is concerned with algorithms which learn from large data sets. For example, the National Survey of Children’s Health (NSCH) is a survey that results in a large data set that can be used with machine learning – can we predict if the child has autism, based on the other survey responses? How accurately can we predict? And what other survey responses are most useful for prediction? In this tutorial, I will show how machine learning can be used to answer these questions.
See also code https://github.com/vas235/ASG3-machine-learning-prep from Vince which treats more than two years, and standardizes some variables between the years, using a JSON config file.
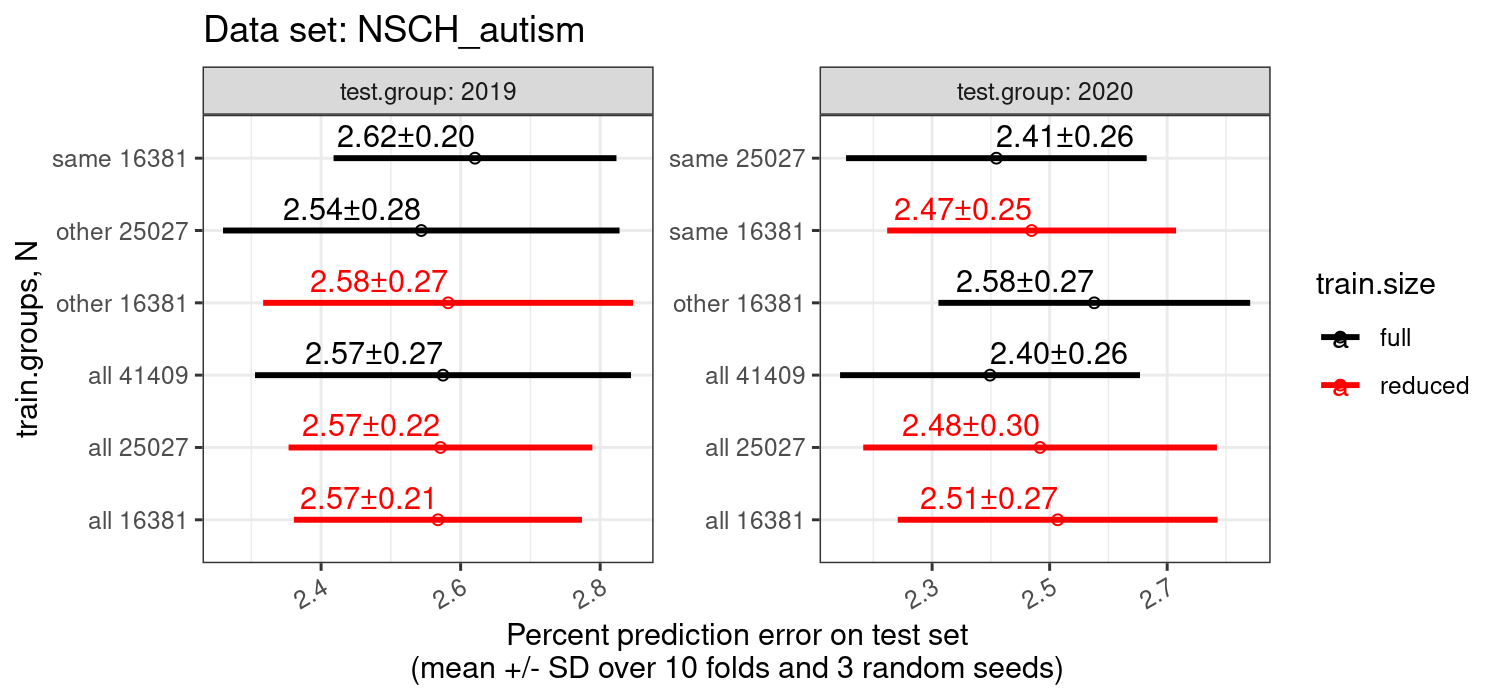
figures-same-other/ contains CSV and figures to show that it is not just size that matters.
HOCKING-slides-2024-02-26-ml-for-autism.tex makes HOCKING-slides-2024-02-26-ml-for-autism.pdf slides with new drawings
makes
drawing-cv-feature-sets.pdf
makes
drawing-cv-same-other-years-1.pdf
drawing-cv-same-other-years-2.pdf
drawing-cv-same-other-years-3.pdf
drawing-cv-same-other-years-4.pdf
download-nsch-mlr3batchmark.R launches jobs, here is a preliminary analysis of how much time and memory they take:
> usage.wide[order(megabytes_max), .(learner_id, task_id, megabytes_min, megabytes_median, megabytes_max, megabytes_length)]
learner_id task_id megabytes_min megabytes_median megabytes_max megabytes_length
<char> <char> <num> <num> <num> <int>
1: classif.cv_glmnet behavior.15 0.0000 0.0000 0.0000 60
2: classif.cv_glmnet comorbidity.30 0.0000 0.0000 0.0000 60
3: classif.cv_glmnet culture.14 0.0000 0.0000 0.0000 60
4: classif.featureless comorbidity.30 0.0000 0.0000 0.0000 60
5: classif.featureless healthcare.88 0.0000 0.0000 0.0000 60
6: classif.rpart birth.24 0.0000 0.0000 0.0000 60
7: classif.rpart comorbidity.30 0.0000 0.0000 0.0000 60
8: classif.rpart culture.14 0.0000 0.0000 0.0000 60
9: classif.rpart healthcare.88 0.0000 0.0000 0.0000 60
10: classif.featureless culture.14 0.0000 0.0000 184.3555 60
11: classif.featureless birth.24 0.0000 0.0000 185.0703 60
12: classif.rpart behavior.15 0.0000 0.0000 195.0234 60
13: classif.featureless behavior.15 0.0000 0.0000 196.5000 60
14: classif.cv_glmnet birth.24 0.0000 0.0000 419.1250 60
15: classif.xgboost culture.14 410.0664 425.7168 516.3867 60
16: classif.xgboost birth.24 411.4688 446.2695 518.8477 60
17: classif.xgboost behavior.15 413.1992 431.9512 519.3633 60
18: classif.xgboost comorbidity.30 411.9727 451.4375 520.8359 60
19: classif.nearest_neighbors culture.14 405.4688 465.7988 531.1367 60
20: classif.nearest_neighbors behavior.15 401.6992 462.6016 552.0781 60
21: classif.nearest_neighbors birth.24 409.3086 472.2266 588.5117 60
22: classif.nearest_neighbors comorbidity.30 435.0664 480.6035 594.1562 60
23: classif.cv_glmnet healthcare.88 0.0000 453.3457 606.5117 60
24: classif.xgboost healthcare.88 519.7617 614.1836 747.3711 60
25: classif.nearest_neighbors healthcare.88 536.2422 613.3730 843.5859 60
26: classif.ranger healthcare.88 1192.5625 1192.5625 1192.5625 1
27: classif.ranger comorbidity.30 1201.4414 1347.5469 1944.3164 30
28: classif.ranger culture.14 898.6367 1336.7637 1966.7070 60
29: classif.ranger behavior.15 1003.0703 1372.0977 2167.9062 60
30: classif.ranger birth.24 1244.2656 1758.0156 2780.9922 43
learner_id task_id megabytes_min megabytes_median megabytes_max megabytes_length
> usage.wide[order(hours_max), .(learner_id, task_id, hours_min, hours_median, hours_max, hours_length)]
learner_id task_id hours_min hours_median hours_max hours_length
<char> <char> <num> <num> <num> <int>
1: classif.featureless culture.14 0.0005555556 0.0008333333 0.001111111 60
2: classif.rpart culture.14 0.0005555556 0.0008333333 0.001111111 60
3: classif.featureless behavior.15 0.0005555556 0.0011111111 0.001388889 60
4: classif.featureless birth.24 0.0005555556 0.0008333333 0.001388889 60
5: classif.rpart comorbidity.30 0.0008333333 0.0008333333 0.001388889 60
6: classif.rpart behavior.15 0.0008333333 0.0011111111 0.001666667 60
7: classif.rpart birth.24 0.0005555556 0.0008333333 0.001666667 60
8: classif.featureless comorbidity.30 0.0005555556 0.0011111111 0.001944444 60
9: classif.featureless healthcare.88 0.0005555556 0.0009722222 0.001944444 60
10: classif.rpart healthcare.88 0.0008333333 0.0011111111 0.002222222 60
11: classif.cv_glmnet culture.14 0.0011111111 0.0016666667 0.002500000 60
12: classif.cv_glmnet behavior.15 0.0019444444 0.0025000000 0.003333333 60
13: classif.cv_glmnet birth.24 0.0013888889 0.0019444444 0.004722222 60
14: classif.cv_glmnet comorbidity.30 0.0016666667 0.0027777778 0.005000000 60
15: classif.cv_glmnet healthcare.88 0.0047222222 0.0094444444 0.020000000 60
16: classif.xgboost culture.14 0.0102777778 0.0166666667 0.027777778 60
17: classif.xgboost behavior.15 0.0169444444 0.0254166667 0.048888889 60
18: classif.xgboost comorbidity.30 0.0252777778 0.0477777778 0.080833333 60
19: classif.nearest_neighbors behavior.15 0.0138888889 0.0291666667 0.084722222 60
20: classif.xgboost birth.24 0.0241666667 0.0366666667 0.087222222 60
21: classif.nearest_neighbors culture.14 0.0122222222 0.0268055556 0.096666667 60
22: classif.nearest_neighbors birth.24 0.0150000000 0.0306944444 0.099444444 60
23: classif.nearest_neighbors comorbidity.30 0.0183333333 0.0398611111 0.170277778 60
24: classif.xgboost healthcare.88 0.0608333333 0.1200000000 0.213333333 60
25: classif.nearest_neighbors healthcare.88 0.0566666667 0.1898611111 0.798888889 60
26: classif.ranger healthcare.88 5.3941666667 5.3941666667 5.394166667 1
27: classif.ranger culture.14 1.1869444444 2.5109722222 6.713055556 60
28: classif.ranger behavior.15 1.5277777778 3.2013888889 8.618611111 60
29: classif.ranger comorbidity.30 3.6255555556 4.6951388889 10.774444444 30
30: classif.ranger birth.24 2.4188888889 5.0616666667 12.538888889 43
learner_id task_id hours_min hours_median hours_max hours_lengthLooks like ranger is by far the slowest and more memory intensive, so for now I will omit that.
Below we see that total time for CV experiment with 2700 iterations is 240 hours, so since we did this in a 4 hour time limit, this is about 60x speedup.
2700: 3.194722222 1810.023 classif.nearest_neighbors all.364
> sum(usage.long$hours)
[1] 240.7103
> sum(usage.long$hours)/4
[1] 60.17757download-nsch-convert-do.R makes download-nsch-convert-do-2019-2020.csv
> out.dt[, table(survey_year, Autism)]
Autism
survey_year Yes No
2019 859 28003
2020 1255 40826download-nsch-counts.R separated out from download-nsch.R
https://docs.google.com/spreadsheets/d/19Tm75T4wNN4yITlXuUMNVc22yzHmmzVcMY1GBVGsEnQ/edit#gid=0 is the source file for NSCH_categories.csv
download-nsch.R makes download-nsch-nrow-ncol.csv and download-nsch-column-counts.csv and NSCH_categories_NA_counts.csv after which I manually added different categories for the least missing columns, NSCH_categories_NA_counts_TDH.csv