- This repository aims to provide scripts that create sysimage that accelerates initialization of IJulia kernel.
- It also will reduce latency when working locally with packages that has a high startup time e.g. Plots.jl or StatsPlots.jl .
$ git clone https://github.com/terasakisatoshi/sysimage_creator.git
$ pip install jupyter jupytext nbconvert ipykernel
$ cd sysimage_creator && make && jupyter notebookThen open your jupyter notebook and select a kernel named Julia-sys 1.6.3
- Almost people can skip this step because your are trying to improve your Jupyter IJulia.jl, to You know what to do.
- Install Julia and Python
- Let's install
jupyter(orjupyterlabinstead) andjupytextvia:
$ pip install jupyter jupytext
$ pip install nbconvert ipykernel cython # I'm not sure, but you will need on Windows.- After that, let's moving on to Step2!!!
- All right. Welcome to Julia! First of all please install Julia from here. Please see platform specific instructions for further installation instructions and if you have trouble installing Julia. You'll learn how to add a path of a program for Julia. Open your terminal and type
juliacommand.
$ julia
_
_ _ _(_)_ | Documentation: https://docs.julialang.org
(_) | (_) (_) |
_ _ _| |_ __ _ | Type "?" for help, "]?" for Pkg help.
| | | | | | |/ _` | |
| | |_| | | | (_| | | Version 1.6.3 (2021-09-23)
_/ |\__'_|_|_|\__'_| | Official https://julialang.org/ release
|__/ |
julia> # congrats!
julia> exit() # exit from julia REPL
$ julia # you can start again.- Moving on to the next step. Let's install PyCall and IJulia and then your environment can initialize jupyter notebook
julia> ENV["PYTHON"]=""; ENV["JUPYTER"]=""
julia> using Pkg; Pkg.add(["PyCall", "IJulia", "Conda"])
julia> using Conda; Conda.add(["jupyter", "jupytext"], channel="conda-forge")
- Tips: you don't have to copy code above line by line. Copy the whole 3 lines. Don't hesitate to include prompt
julia>. Then just do paste to your julia REPL. - Tips: Setting
ENV["PYTHON"]=""; ENV["JUPYTER"]=""makes julia select Python provided via Conad.jl. Conda.jl uses the miniconda Python environment, which only includes conda and its dependencies. If you have LITTLE experience with Python, just follow the instructions above is fine. - Tips: Ah, you've remembered you can use Python and Jupyter from your termina? e.g.
$ python
>>>$ pip install numpyor whatever
$ jupyter notebookIf so, you may consider do the following command:
julia> run(`pip install jupyter jupytext nbconvert ipykernel`)
julia> ENV["PYTHON"]=Sys.which("python3"); ENV["JUPYTER"]=Sys.which("jupyter")
julia> using Pkg; Pkg.build(["PyCall", "IJulia"])What does it do? Well, read the code: deps/build.jl.
Anyway, after finished the installation, let's initialize Juptyer notebook:
julia> using IJulia; notebook(dir=pwd())- Ah... also make sure you can use
$ makecommand in your terminal. - Just run
make. It will make sysimage namedsys.{DLEXT}, whereDLEXTisdylib, dll or so.
$ make
...
...
...
$ ls
sys.dylib # e.g. macOS users
- You can feel how much time we can reduce the latency of loading Julia packages when we use our sysimage:
$ cat benchmark.jl
# use standard julia
@time run(`jupytext --to ipynb --execute testout_naive.jl`)
# use sys.${DLEXT} as sysimage
@time run(`jupytext --to ipynb --execute testout_sys.jl`)
$ julia --project=@. benchmark.jlor just run make test :D.
- Open Jupyter Notebook as always:
$ jupyter notebook- Then you'll see New kernel for Julia named
Julia-sys 1.6.3.
- Please select it. This kernel is generated by
installkernel.jlwhich is stored in this repository.
$ cat ~/Library/Jupyter/kernels/julia-sys-1.6/kernel.json
{
"display_name": "Julia-sys 1.6.3",
"argv": [
"/Applications/Julia-1.6.app/Contents/Resources/julia/bin/julia",
"-i",
"--color=yes",
"--project=@.",
"--sysimage=sys", # <--- this `sys` is a name of sysimage
"/Users/<your-user-name>/.julia/packages/IJulia/e8kqU/src/kernel.jl",
"{connection_file}"
],
"language": "julia",
"env": {},
"interrupt_mode": "signal"
}- If you click
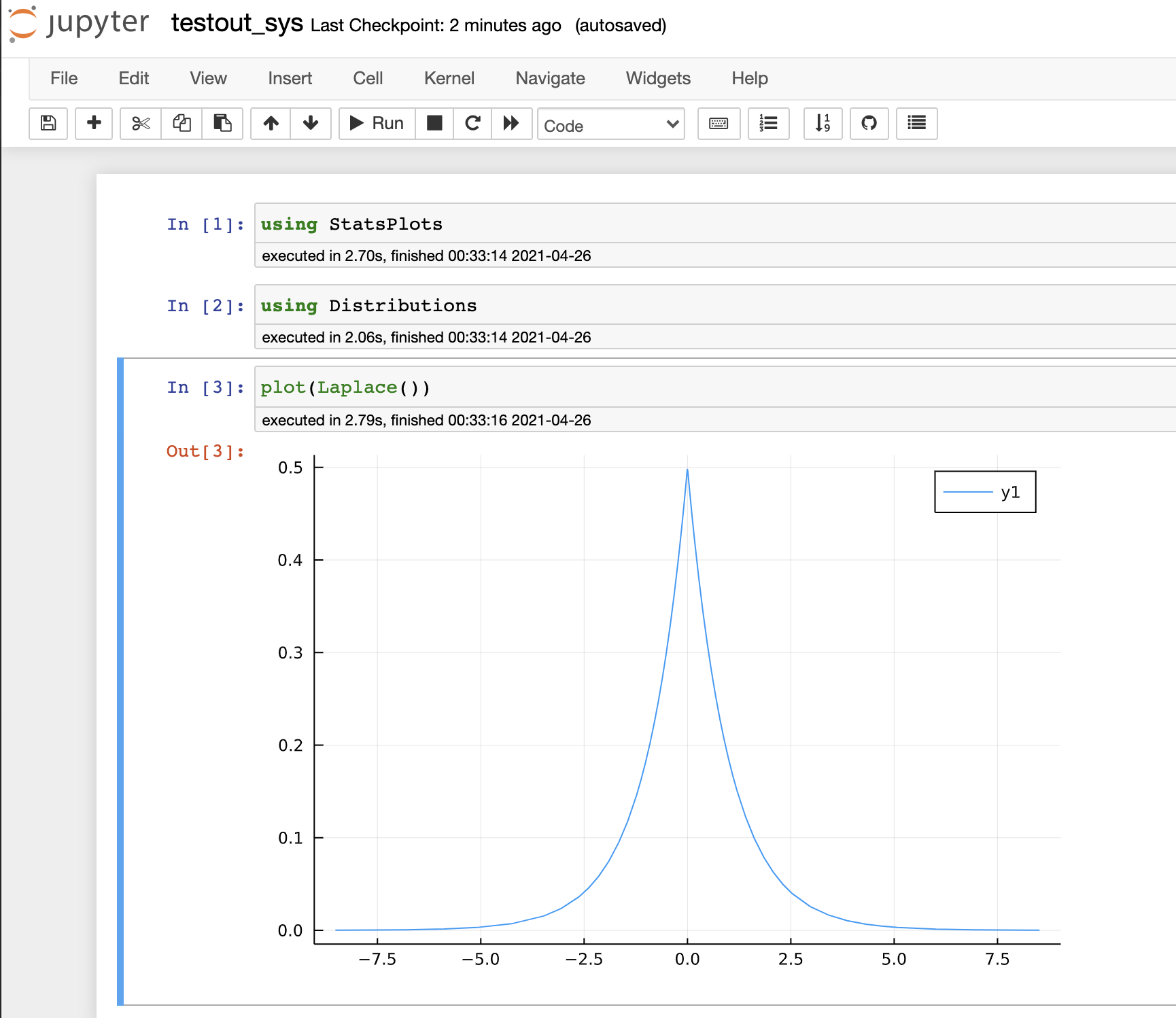
testout_sys.jlfile, Jupyter recognize this file as notebook, and initialize it with option--sysimage=sys. - Here is a result of running
testout_sys.jlusing our sysimage:
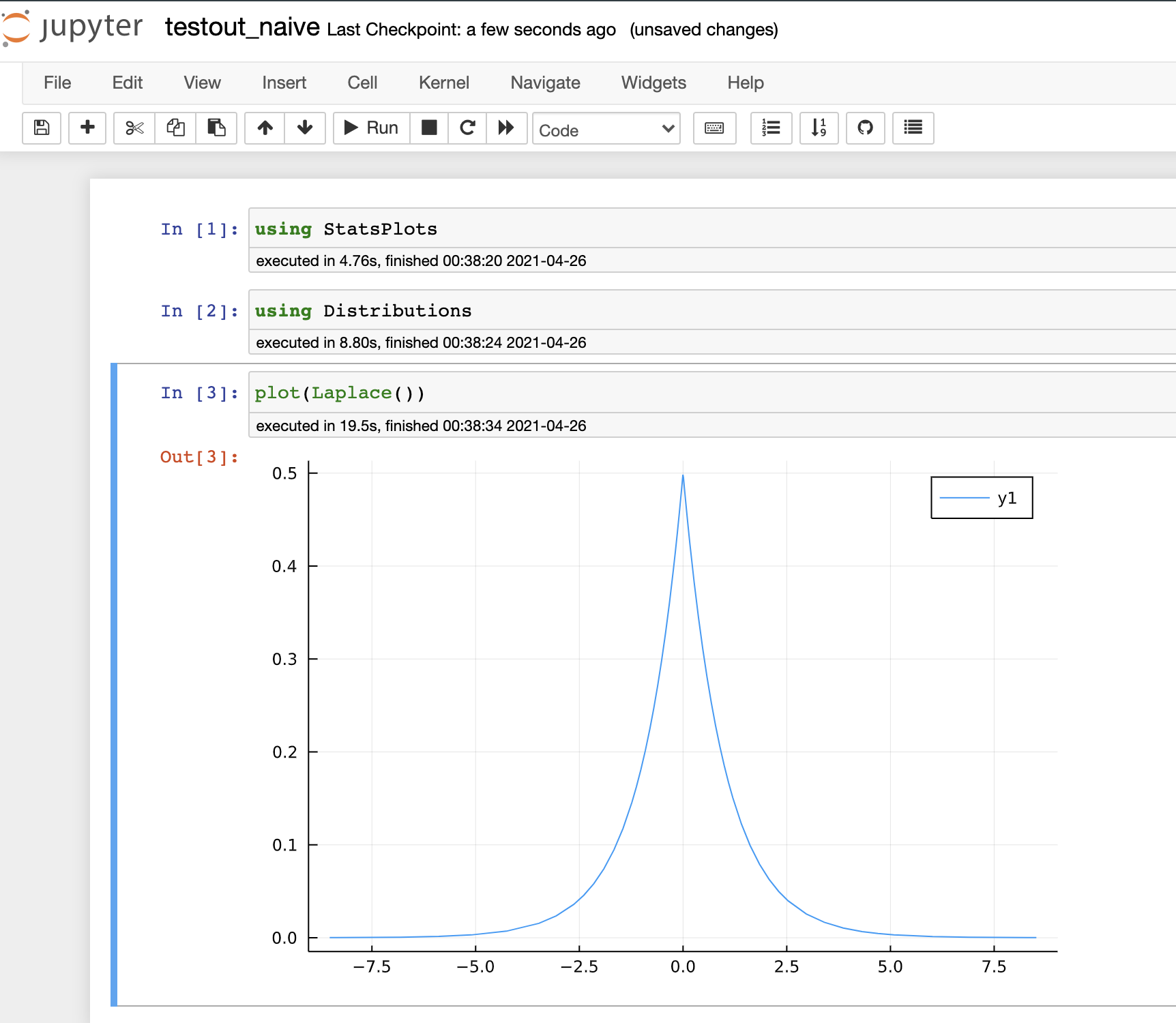
- Here is a result of running
testout_naive.jlusing standard sysimage, which is so slow: